Comparative Efficiency of Plant Genetic Transformation Methods: A Guide for Researchers and Developers
This article provides a comprehensive analysis of the efficiency, applications, and future trajectories of modern plant genetic transformation techniques.
Comparative Efficiency of Plant Genetic Transformation Methods: A Guide for Researchers and Developers
Abstract
This article provides a comprehensive analysis of the efficiency, applications, and future trajectories of modern plant genetic transformation techniques. Tailored for researchers, scientists, and biotechnology professionals, it systematically compares established methods like Agrobacterium-mediated transformation and biolistics with emerging tissue culture-free strategies. We explore the foundational principles of plant regeneration, delve into specific methodological protocols, and address key bottlenecks through advanced optimization strategies such as developmental regulators and improved gene gun designs. A critical validation and comparative analysis section equips the reader to select the most efficient method for their specific project, with conclusions highlighting the transformative potential of these technologies for accelerating crop improvement and biopharmaceutical production.
The Bedrock of Plant Genetic Engineering: Core Principles and Regeneration Biology
Defining Plant Genetic Transformation and Its Role in Crop Improvement
Plant genetic transformation is a foundational biotechnology that enables the precise introduction of genes with known functional characteristics—such as high yield potential, stress resistance, disease and pest tolerance, and enhanced nutritional profiles—into target plants [1]. This process allows recipient plants to acquire novel agricultural traits while preserving their original genetic foundations, representing a significant advancement over traditional breeding methods [1]. Since the first genetically modified crop was successfully generated in 1983, researchers have developed over 200 transgenic plants spanning 35 botanical families, including major crops like rice, corn, wheat, tomatoes, cotton, and soybeans [1]. The global significance of this technology is reflected in its market trajectory, valued at $1.58 billion in 2024 and projected to reach $3.03 billion by 2035, demonstrating its expanding role in addressing agricultural challenges [2].
The advancement of plant regeneration technology is critical for addressing complex and dynamic climate challenges, ultimately ensuring global agricultural sustainability [3]. Genetic transformation technologies have become indispensable tools for both fundamental research and applied crop improvement, enabling scientists to elucidate gene functions and rapidly develop varieties with enhanced agronomic traits [1] [4]. As the global population continues to grow and climate change introduces new production constraints, these technologies offer promising pathways to develop crops with improved resilience, productivity, and nutritional quality [5].
Fundamental Principles and Methodologies of Plant Genetic Transformation
Classification of Transformation Methods
Plant genetic transformation methodologies can be broadly classified into two primary categories: direct gene transfer methods and bio-mediated transformation methods [1]. Direct gene transfer includes techniques such as microprojectile bombardment (gene gun), protoplast methods, liposome-mediated transfer, the pollen tube pathway, electroshock conversion, and PEG-mediated transformation [1]. Among these, microprojectile bombardment represents the most prominent direct transfer method, where DNA-coated metal particles are physically propelled into plant cells [4]. This approach is particularly valuable for species and genotypes that are less susceptible to biological transformation methods, though it often results in higher transgene copy numbers and increased instances of gene rearrangement [4].
Bio-mediated transformation predominantly utilizes biological vectors such as Agrobacterium tumefaciens and plant viruses to facilitate gene transfer [1]. Agrobacterium-mediated transformation has emerged as the preferred method for many plant species due to its simplicity, low cost, high transformation efficiency, and tendency to produce transgenic plants with lower transgene copy numbers and greater stability [1] [6]. This natural gene transfer mechanism exploits the soil bacterium's ability to transfer a segment of its DNA (T-DNA) into the host plant genome, a process that has been refined and optimized for laboratory transformation over several decades [1] [4].
The Centrality of Plant Regeneration
A fundamental prerequisite for successful genetic transformation is the establishment of a high-frequency plant regeneration system [1]. Regeneration describes the process by which plant tissues or organs repair and replace themselves following damage or environmental stress, a remarkable capability that finds extensive application in agricultural and horticultural techniques including cutting propagation, grafting, and tissue culture methodologies [1]. The concepts of cellular totipotency and pluripotency serve as the cytological foundation of plant regeneration, wherein somatic plant cells can be reprogrammed through hormonal interactions to develop into independent plants or organs via cell division and differentiation [1].
Higher plants regenerate through meristematic tissues, primarily the shoot apical meristem (SAM), root apical meristem (RAM), and lateral meristems [1]. These meristematic stem cells maintain their population through cell division while simultaneously differentiating into diverse tissue and organ cell types, representing the fundamental production source for complex biological structures in higher plants [1]. Contemporary research has identified specific classes of genes that actively promote plant regeneration during transformation processes, enabling the development of plant materials with heightened genetic transformation efficiency [1].
Table 1: Comparison of Major Plant Genetic Transformation Techniques
| Method | Key Features | Advantages | Limitations | Primary Applications |
|---|---|---|---|---|
| Agrobacterium-mediated | Uses A. tumefaciens to transfer T-DNA to plant cells [1] | Simple, low cost, high efficiency, low transgene copy number [6] | Genotype-dependent, low efficiency in some species [7] | Dicot transformation, stable transgene integration [1] |
| Particle Bombardment | DNA-coated metal particles propelled into plant cells [4] | Species-independent, no vector requirements [4] | High equipment cost, complex integration patterns [4] | Monocot transformation, organelle transformation [4] |
| Pollen-tube Pathway | Exogenous DNA introduced via pollen tube after pollination [1] | Avoids tissue culture, technically simple [1] | Low efficiency, limited reproducibility [1] | Cotton, melon, soybean transformation [1] |
| Protoplast Transformation | Direct DNA uptake by plant cells without cell walls [1] | High transformation frequency, synchronous treatment [1] | Difficult regeneration, genotype limitations [1] | Transient expression studies, single-cell analysis [1] |
Comparative Analysis of Major Transformation Methods
Agrobacterium-mediated Transformation
Agrobacterium-mediated transformation has become the most widely used method for plant genetic engineering, with approximately 85% of transgenic plants obtained using this approach [4]. The process begins with the recognition of plant signals by Agrobacterium, followed by bacterial attachment to wounded plant tissue and the subsequent transfer of T-DNA from the bacterium to the host plant genome [6]. Successful infection depends on both plant genotypes and Agrobacterium strains, with significant variation observed among different species and varieties [6].
The efficiency of Agrobacterium-mediated transformation is influenced by numerous factors, including explant type, Agrobacterium concentration, co-cultivation time, and medium composition [4]. Optimization studies have demonstrated that collecting Agrobacterium at a concentration of OD₆₅₀ = 0.6 and using a suspension medium containing dithiothreitol to infect half-seed cotyledonary explants can achieve infection efficiencies exceeding 96% [4]. Furthermore, the addition of phenolic compounds like acetosyringone has been found essential for induction of the virulence genes, while antioxidant reagents such as L-cysteine and dithiothreitol improve T-DNA delivery by inhibiting the activity of plant pathogen-response and wound-response enzymes [6].
Recent innovations have focused on enhancing Agrobacterium-mediated transformation through the use of auxiliary solutions. One study demonstrated that an Agrobacterium Auxiliary Solution (AAS) containing Silwet L-77 and hormone mixtures significantly improved hairy root transformation rates in soybean [7]. This combination increased total root and cotyledon transformation efficiencies compared to control treatments, highlighting the importance of chemical enhancers in optimizing transformation protocols [7].
Particle Bombardment and Direct DNA Transfer Methods
Particle bombardment, also known as biolistics or the gene gun approach, represents a physically-mediated transformation method that involves coating micron-sized gold or tungsten particles with DNA and accelerating them into plant cells using high-pressure helium gas [4]. This technique offers the significant advantage of being less constrained by plant genotype or species barriers compared to Agrobacterium-mediated methods, making it particularly valuable for transforming cereals and other species that have historically shown resistance to Agrobacterium infection [4].
The particle bombardment method does not require vector-specific sequences for DNA integration and enables the transformation of organelles, including chloroplasts and mitochondria, which is more challenging with biological vectors [4]. However, this approach typically results in more complex integration patterns, including higher transgene copy numbers and increased instances of gene rearrangement [4]. The equipment costs are substantially higher than Agrobacterium-based methods, and the process often produces more transgenic plants with silencing or unstable expression of the introduced genes due to complex integration loci [4].
Emerging and Specialized Transformation Techniques
Pollen-tube pathway transformation represents a unique direct DNA transfer method that utilizes the natural pathway of pollen tube growth to introduce exogenous genes into fertilized embryos [1]. First demonstrated in the 1970s, this technique involves injecting foreign DNA into the ovary after pollination, allowing the pollen tube to serve as a conduit for the DNA to enter the fertilized egg cell [1]. The major advantage of this approach is its bypass of the complex tissue culture process, making the technology relatively simple and accessible without requiring sophisticated laboratory facilities [1]. This method has been successfully applied to important crops including cotton, melon, soybean, and wheat, with transformation efficiencies reaching up to 2.54% in optimal conditions [1].
More recently, nanoparticle-mediated transformation has emerged as a promising alternative to conventional methods, though it was not specifically detailed in the search results. Similarly, floral dip methods, widely used in Arabidopsis transformation, represent another Agrobacterium-based approach that avoids tissue culture by directly infecting developing flowers.
Table 2: Quantitative Comparison of Transformation Efficiencies Across Methods and Species
| Plant Species | Transformation Method | Efficiency Range (%) | Key Factors Influencing Efficiency | Optimal Explant Type |
|---|---|---|---|---|
| Soybean [6] [4] | Agrobacterium-mediated | 0.21-10% | Genotype, GA and ZR levels, MeJA content [6] | Cotyledonary nodes [4] |
| Soybean [7] | Hairy root (A. rhizogenes) | 30-60% (cotyledon), up to 80% (stem) | Auxiliary solution, vector size [7] | Cotyledons, hypocotyls [7] |
| Rice [4] | Agrobacterium-mediated | Up to 23% | Genotype, selection system [4] | Immature embryos [4] |
| Maize [4] | Agrobacterium-mediated | 30-40% | Genotype, embryo quality [4] | Immature embryos [4] |
| Paphiopedilum Maudiae [1] | Pollen-tube pathway | 2.54% | Developmental stage, DNA delivery timing [1] | Ovules [1] |
Experimental Data: Method Efficiencies and Optimization Protocols
Soybean Transformation Efficiency Across Genotypes
Comparative studies have revealed significant variation in transformation efficiency among different soybean genotypes, highlighting the genotype-dependent nature of genetic transformation technologies [6]. Research evaluating ten soybean cultivars demonstrated dramatic differences in transformation efficiency, with high-efficiency genotypes like Williams 82, Shennong 9, and Bert achieving transformation rates of 6.71%, 5.32%, and 5.13% respectively, while low-efficiency genotypes such as General, Liaodou 16, and Kottman showed transformation efficiencies below 1% [6]. These differences in transformability were correlated with specific physiological and molecular factors, including hormone levels, gene expression patterns, and enzyme activities [6].
High-efficiency genotypes exhibited higher gibberellin (GA) levels and increased expression of soybean GA20ox2 transcripts, along with higher zeatin riboside (ZR) content and DNA quantity, and relatively higher expression of soybean IPT5, CYCD3, and CYCA3 genes [6]. Conversely, these genotypes showed lower methyl jasmonate (MeJA) content, polyphenol oxidase (PPO) activity, and peroxidase (POD) activity, along with reduced expression of OPR3, PPO1, and PRX71 genes [6]. These findings suggest that GA and ZR function as positive plant factors for Agrobacterium-mediated soybean transformation by facilitating germination and growth and increasing the number of cells in the DNA synthesis cycle, respectively [6]. Meanwhile, MeJA, PPO, POD, and abscisic acid (ABA) act as negative plant factors by inducing defense reactions and repressing germination and growth [6].
Detailed Experimental Protocol for Agrobacterium-mediated Soybean Transformation
An optimized protocol for Agrobacterium-mediated soybean transformation has been developed through systematic investigation of factors affecting infection and regeneration efficiency [4]. The following represents a detailed methodology for achieving high transformation efficiency in soybean:
Explant Preparation and Bacterial Infection:
- Surface-sterilize mature soybean seeds and imbibe for approximately 24 hours to obtain "half-seed" cotyledonary explants [4].
- Prepare Agrobacterium suspension by collecting bacteria at logarithmic growth phase (OD₆₅₀ = 0.6) and resuspending in infection medium containing 154.2 mg/L dithiothreitol [4].
- Infect explants by immersion in Agrobacterium suspension for 15-30 minutes with gentle agitation [7].
Co-cultivation and Selection:
- Transfer infected explants to co-cultivation medium with their adaxial side facing upward [4].
- Co-cultivate for 5 days in darkness at 22-25°C to allow T-DNA transfer and initial integration [4].
- Transfer explants to shoot induction medium containing appropriate selection agents (e.g., phosphinothricin at 5 mg/L) to inhibit growth of non-transformed tissues [6].
Shoot Elongation and Rooting:
- For shoot elongation, transfer explants with developing shoots to medium containing optimal hormone combinations—specifically 1.0 mg/L GA₃ and 0.1 mg/L IAA—which has been shown to increase elongation rates by 11-18% compared to standard protocols [4].
- Individual elongated shoots are subsequently transferred to rooting medium containing auxins such as IBA or NAA to promote root development [4].
- Finally, established plantlets are acclimatized to greenhouse conditions and grown to maturity for seed production [4].
Molecular Confirmation: Putative transgenic plants are verified through multiple methods including GUS histochemical staining, PCR analysis, herbicide painting (for herbicide resistance genes), and absolute quantification PCR to determine transgene copy number [4].
Successful plant genetic transformation requires specific reagents, vectors, and biological materials optimized for different plant species and transformation methods. The following toolkit summarizes critical components referenced in the search results:
Table 3: Essential Research Reagents for Plant Genetic Transformation
| Reagent Category | Specific Examples | Function & Application | Optimization Notes |
|---|---|---|---|
| Agrobacterium Strains | EHA105, K599, Ar. Qual [7] | T-DNA delivery to plant cells | Strain selection affects host range and efficiency [7] |
| Vector Systems | pAGM4673, pTOPO-Blunt, Ruby vectors [7] | Gene cloning and expression | Smaller vectors (e.g., GFP) show higher efficiency [7] |
| Selection Agents | Phosphinothricin (5 mg/L), Antibiotics [6] | Selection of transformed tissues | Concentration optimization critical for regeneration [6] |
| Phenolic Inducers | Acetosyringone (40 mg/L) [7] | Vir gene induction in Agrobacterium | Essential for transformation competence [6] |
| Hormonal Supplements | GA₃ (1.0 mg/L), IAA (0.1 mg/L), 6-BA [4] [7] | Regulate plant regeneration | Optimal combinations improve shoot elongation [4] |
| Antioxidants | Dithiothreitol (154.2 mg/L), L-cysteine [4] | Suppress plant defense responses | Improve T-DNA delivery [6] |
| Surfactants | Silwet L-77 (100 μL/L) [7] | Enhance tissue penetration | Component of auxiliary solutions [7] |
Current Innovations and Future Perspectives
Advanced Genome Editing Tools
The field of plant genetic transformation is being revolutionized by CRISPR-based genetic editing technologies, which have rapidly become the most widely used tools in genome engineering due to their accuracy, cost-effectiveness, and ease of implementation [8] [2]. Comparative studies have evaluated the efficiency and specificity of Cas nucleases from different bacterial species, including Cas9 from Streptococcus pyogenes (SpCas9) and Staphylococcus aureus (SaCas9), as well as Cas12a from Francisella novicida and Lachnospiraceae bacterium [8]. These studies have found that SaCas9 is comparatively most efficient at inducing mutations, and that "high-fidelity" variants of Cas9 can effectively reduce off-target mutations in plants [8].
Innovations around CRISPR 2.0 and base editing technologies are gaining momentum, ushering in the next wave of biologically engineered products [2]. Base editing enables precise nucleotide changes without creating double-strand breaks in DNA, offering greater precision and potentially higher efficiency for specific applications [2]. Furthermore, the integration of artificial intelligence and machine learning in genomic research is playing an increasingly important role in identifying gene-editing targets, predicting off-target effects, and modeling disease pathways, thereby making crop improvement programs more efficient and less risky [2].
Hormonal Regulation and Signaling Pathways
Recent research has elucidated the critical role of hormonal networks in regulating plant regeneration and transformation efficiency [3] [9]. Studies on bolting and flowering in various plant species, including leaf lettuce and Saposhnikovia divaricata, have demonstrated that plant hormone signal transduction pathways are decisive factors in controlling developmental transitions [10] [9]. In leaf lettuce, silencing of the serine/threonine protein kinase (STPK) gene affected bolting through modifications in auxin (IAA), gibberellin (GA3), and abscisic acid (ABA) contents [10]. Similarly, transcriptome analysis of Saposhnikovia divaricata identified auxin-related genes IAA and TIR1 as key regulators in the bolting and flowering process [9].
These findings highlight the intricate hormonal crosstalk that underlies plant development and regenerative capacity. The antagonistic relationships between hormones—such as ABA acting as an antagonist to GA [6]—directly impact the efficiency of transformation protocols, particularly during the critical stages of shoot induction and elongation [6] [4]. Future transformation protocols will likely incorporate more sophisticated hormonal manipulations based on improved understanding of these signaling networks.
Addressing Current Challenges and Future Directions
Despite significant advancements, plant genetic transformation continues to face several formidable challenges. Genotype-dependent transformation efficiency remains a major constraint, particularly for commercially valuable elite cultivars that are often recalcitrant to standard transformation protocols [6] [7]. Additionally, regulatory scrutiny and ethical concerns often slow the deployment of gene-edited crops, particularly in certain regions with restrictive policies toward genetically modified organisms [2] [5]. The high costs of research and development, coupled with intellectual property complexities, present additional barriers to widespread adoption of transformation technologies [2].
Future research directions will likely focus on developing genotype-independent transformation systems through the identification and utilization of regeneration-promoting genes [1]. The application of single-cell and spatial transcriptome technologies promises to provide unprecedented insights into the molecular foundations of plant regeneration and cellular totipotency [3] [1]. Furthermore, the emergence of synthetic biology approaches combining engineering principles with biological design is opening new frontiers in the creation of plants with novel functions, potentially expanding the capabilities of crop improvement beyond traditional agronomic traits [2] [5].
As these technologies continue to evolve, plant genetic transformation will play an increasingly vital role in global efforts to ensure food security, enhance nutritional quality, and develop sustainable agricultural systems capable of withstanding the challenges of climate change and population growth [3] [5]. The ongoing refinement of transformation methodologies will enable more precise and efficient genetic modifications, accelerating the development of next-generation crops with improved productivity, resilience, and value-added traits.
Cellular potency, the capacity of a single cell to differentiate into various cell types, establishes the fundamental framework for regeneration in multicellular organisms. This potency exists on a spectrum, with totipotency and pluripotency representing its most powerful states. In mammals, totipotency is a transient characteristic of the earliest embryonic cells, such as the zygote and early blastomeres (up to the 4-cell stage in humans), enabling them to generate an entire organism, including both embryonic and extraembryonic tissues like the placenta [11] [12]. Following several cell divisions, these totipotent cells transition into a pluripotent state. Pluripotent cells, found in the inner cell mass (ICM) of the blastocyst, can give rise to all cells of the adult body—derived from the three germ layers (ectoderm, mesoderm, and endoderm)—but cannot form extraembryonic structures [13] [14].
In plants, the concepts of totipotency and pluripotency are equally pivotal, underpinning their remarkable regenerative capabilities. Theoretically, every somatic plant cell is totipotent, retaining the ability to regenerate a complete new plant [15]. This totipotency is the cytological foundation for plant regeneration, which is exploited extensively in agricultural techniques such as cutting propagation and advanced biotechnology like tissue culture and genetic transformation [1]. Pluripotent cells in plants are found in meristematic tissues (e.g., shoot apical meristem, root apical meristem), which are responsible for generating specific lineages of organs and tissues [1] [15].
Understanding the distinctions between these potent states is not merely an academic exercise; it is crucial for advancing regenerative medicine and improving plant genetic transformation methodologies. This review objectively compares the developmental potential, molecular signatures, and experimental applications of totipotent and pluripotent cells, with a specific focus on their role as the biological basis for regeneration.
Table 1: Core Definitions and Key Characteristics of Potency States
| Feature | Totipotency | Pluripotency |
|---|---|---|
| Definition | Ability to form a complete organism plus extraembryonic tissues [11] | Ability to form all cells of the three germ layers, but not extraembryonic tissues [11] |
| Natural Occurrence (Mammals) | Zygote, early blastomeres (e.g., 2- to 4-cell stage) [12] | Inner cell mass (ICM) of the blastocyst [13] |
| Natural Occurrence (Plants) | Somatic cells under specific induction [15] | Cells in shoot/root apical meristems [1] |
| Key In Vitro Models (Mammals) | Totipotent-like cells (e.g., 2-cell-like cells in mouse) [16] | Embryonic Stem Cells (ESCs), Induced Pluripotent Stem Cells (iPSCs) [11] [14] |
| Key In Vitro Models (Plants) | Embryogenic callus in somatic embryogenesis [15] | Pluripotent callus in de novo organogenesis [15] |
Molecular Hallmarks and Functional Assays
The distinct developmental capacities of totipotent and pluripotent cells are governed by unique molecular landscapes. A core network of transcription factors, including OCT4, SOX2, and NANOG, is essential for maintaining pluripotency [13] [12]. These factors regulate a suite of genes that keep the cell in an undifferentiated state, poised for lineage specification. Totipotent cells, by contrast, exhibit a more open chromatin state and express a distinct set of genes, such as Zscan4 and Eomes, which are associated with their expanded developmental potential [11]. The transition from totipotency to pluripotency involves significant epigenetic reprogramming, including DNA demethylation and specific histone modifications, which progressively restrict cell fate [11] [13].
Functionally, these states are validated through rigorous in vivo and in vitro assays. The gold-standard functional test for totipotency is the ability of a single cell to generate a complete, fertile organism, including all extraembryonic tissues, as demonstrated by tetraploid complementation in mice [13]. For pluripotent cells, key assays include teratoma formation—where injected cells form tumors containing tissues from all three germ layers—and chimera formation, where the cells contribute to various tissues in a developing host embryo [11] [13].
Figure 1: Developmental Trajectory from a Totipotent Zygote. The totipotent zygote gives rise to all embryonic (via the pluripotent ICM) and extraembryonic tissues (via the trophectoderm).
Comparative Efficiency in Plant Genetic Transformation
In plant biotechnology, the principles of totipotency and pluripotency are directly harnessed for genetic transformation. The efficiency of these processes is heavily dependent on the plant's regenerative capacity, which is often the primary bottleneck [1] [17]. Transformation strategies can be broadly categorized into those relying on somatic embryogenesis (involving a totipotent state) and de novo organogenesis (involving a pluripotent state) [15].
- Somatic Embryogenesis: This process involves reprogramming a somatic cell to become a totipotent embryogenic callus cell, which can then develop into a somatic embryo and a whole plant. It can be direct or, more commonly, indirect via a callus phase [15]. This pathway is central to many transformation protocols for major crops like maize and rice.
- De novo Organogenesis: This pathway involves the formation of adventitious roots or shoots directly from an explant or indirectly through a pluripotent callus. This callus is not embryogenic but can be induced to form organ primordia. It reflects the pluripotency of certain cell types and is widely used in plant tissue culture [15].
The efficiency of these regeneration pathways is quantitatively measured by key metrics, as summarized in Table 2. The choice of pathway and explant has a direct impact on transformation success, timeframes, and the risk of undesirable genetic variations.
Table 2: Comparative Analysis of Plant Regeneration Pathways for Genetic Transformation
| Regeneration Pathway | Cellular Basis | Typical Explants | Transformation Efficiency Range | Key Advantages | Key Limitations |
|---|---|---|---|---|---|
| Somatic Embryogenesis | Totipotency [15] | Immature embryos, shoot tips [15] | Highly variable; e.g., 2.7% to 45.3% in wheat [17] | High multiplication rate; produces many plantlets [15] | Long duration; high somaclonal variation risk [15] |
| De novo Organogenesis | Pluripotency [15] | Leaves, roots, floral tissues [15] | High in model plants (e.g., Arabidopsis, tomato); low in cereals [17] | Direct regeneration avoids callus phase; lower somaclonal variation [15] | Fewer regenerated plantlets; strong genotype dependence [17] [15] |
Experimental Protocols and Enhancement Strategies
Protocol for Agrobacterium-Mediated Transformation via Somatic Embryogenesis
This is a standard protocol for generating transgenic plants in cereals like maize, relying on indirect somatic embryogenesis [15] [18].
- Explant Preparation: Sterilize immature embryos (1.0-2.0 mm in size) harvested from plants.
- Callus Induction (Dedifferentiation): Culture embryos on a Callus-Inducing Medium (CIM). This medium is rich in auxin (e.g., 2,4-D) and has low cytokinin, promoting the formation of a totipotent, embryogenic callus. Incubate in the dark for 2-4 weeks.
- Agrobacterium Co-cultivation: Harvest the embryogenic callus and immerse in a suspension of Agrobacterium tumefaciens carrying the gene of interest. Co-cultivate for a short period to allow T-DNA transfer.
- Selection and Regeneration (Differentiation): Transfer the callus to a fresh CIM containing antibiotics to eliminate Agrobacterium and select for transformed cells (using a selectable marker like an antibiotic/herbicide resistance gene). After selection, transfer the transgenic callus to a Shoot-Inducing Medium (SIM), which has high cytokinin and low auxin, to promote shoot formation.
- Rooting and Acclimatization: Excise developed shoots and transfer to a Root-Inducing Medium (RIM) containing auxin to encourage root development. Finally, transfer the plantlets to soil for acclimatization.
Enhancing Efficiency with Developmental Regulators
A major advancement in overcoming genotype-dependent regeneration is the use of developmental regulators (DRs). These are transcription factors that can enhance or induce the formation of totipotent or pluripotent cells [17] [18]. Their application can dramatically improve transformation efficiency, as shown by experimental data.
Table 3: Quantitative Impact of Key Developmental Regulators on Transformation Efficiency
| Developmental Regulator | Target Species | Function | Experimental Impact on Efficiency |
|---|---|---|---|
| TaWOX5 [17] | Wheat (Triticum aestivum) | Promotes pluripotency acquisition in callus; shoot regeneration. | Increased transformation efficiency in recalcitrant variety 'Jimai22' from 5.8% to 55.4% [17]. |
| ZmBBM & ZmWUS2 [17] | Maize (Zea mays) | Activates cell proliferation and somatic embryogenesis. | Combination enhanced transformation in monocots; enabled direct transformation of mature seed-derived tissues, bypassing callus culture [17]. |
| GRF4-GIF1 [18] | Wheat (Triticum aestivum) | Promotes cell proliferation and shoot regeneration. | Increased regeneration frequency in hexaploid wheat from 12.7% to 61.8% [18]. |
| REF1 [18] | Tomato (Solanum lycopersicum) | Wound-signaling molecule promoting callus and bud regeneration. | Increased regeneration efficiency by 5- to 19-fold and transformation efficiency by 6- to 12-fold in wild tomato [18]. |
Figure 2: Workflow for Enhanced Plant Genetic Transformation. Key steps show how developmental regulators (DRs) enhance the formation of totipotent callus.
The Scientist's Toolkit: Key Research Reagents
Leveraging totipotency and pluripotency in research requires a specific set of reagents and tools. The following table details essential materials used in this field.
Table 4: Essential Research Reagents for Studying and Applying Totipotency/Pluripotency
| Research Reagent / Tool | Category | Primary Function in Research |
|---|---|---|
| Yamanaka Factors (OCT4, SOX2, KLF4, c-MYC) [11] | Reprogramming Factors | Reprogram somatic cells into induced pluripotent stem cells (iPSCs) [11]. |
| 2i/LIF Culture System [13] | Cell Culture Medium | Maintains mouse embryonic stem cells (mESCs) in a naive pluripotent state by inhibiting differentiation signals [13]. |
| Morphogenic Regulators (BBM, WUS, WOX) [17] [18] | Developmental Regulators | Enhances plant transformation efficiency by promoting callus formation and shoot regeneration in recalcitrant species [17]. |
| Agrobacterium tumefaciens Strain (e.g., EHA105, GV3101) [1] [19] | Transformation Vector | Delivers foreign DNA (T-DNA) into plant cells for stable genetic transformation [1]. |
| Callus-Inducing Medium (CIM) [15] | Plant Tissue Culture Medium | Induces dedifferentiation of somatic plant cells to form totipotent callus, typically with high auxin (2,4-D) [15]. |
| Anti-OCT4 / Anti-SOX2 / Anti-NANOG Antibodies [12] | Molecular Biology Reagents | Detects and validates the presence of core pluripotency factors via immunostaining or Western blot. |
The objective comparison between totipotent and pluripotent states reveals a clear hierarchy of developmental potential, each with distinct molecular signatures and functional applications. In mammalian systems, pluripotent stem cells like ESCs and iPSCs are currently more therapeutically accessible, while research into totipotent-like cells deepens our understanding of early development. In plant science, both states are directly exploited, with somatic embryogenesis (totipotency) and de novo organogenesis (pluripotency) serving as the two pillars of in vitro regeneration and genetic transformation.
The efficiency of these processes, particularly in plants, is no longer solely dependent on innate genotype. The strategic application of developmental regulators is demonstrably overcoming traditional bottlenecks, as evidenced by the quantitative data on factors like TaWOX5 and BBM/WUS. This progress underscores that the future of regenerative biology and crop engineering lies in the precise manipulation of the very molecular pathways that govern cellular totipotency and pluripotency.
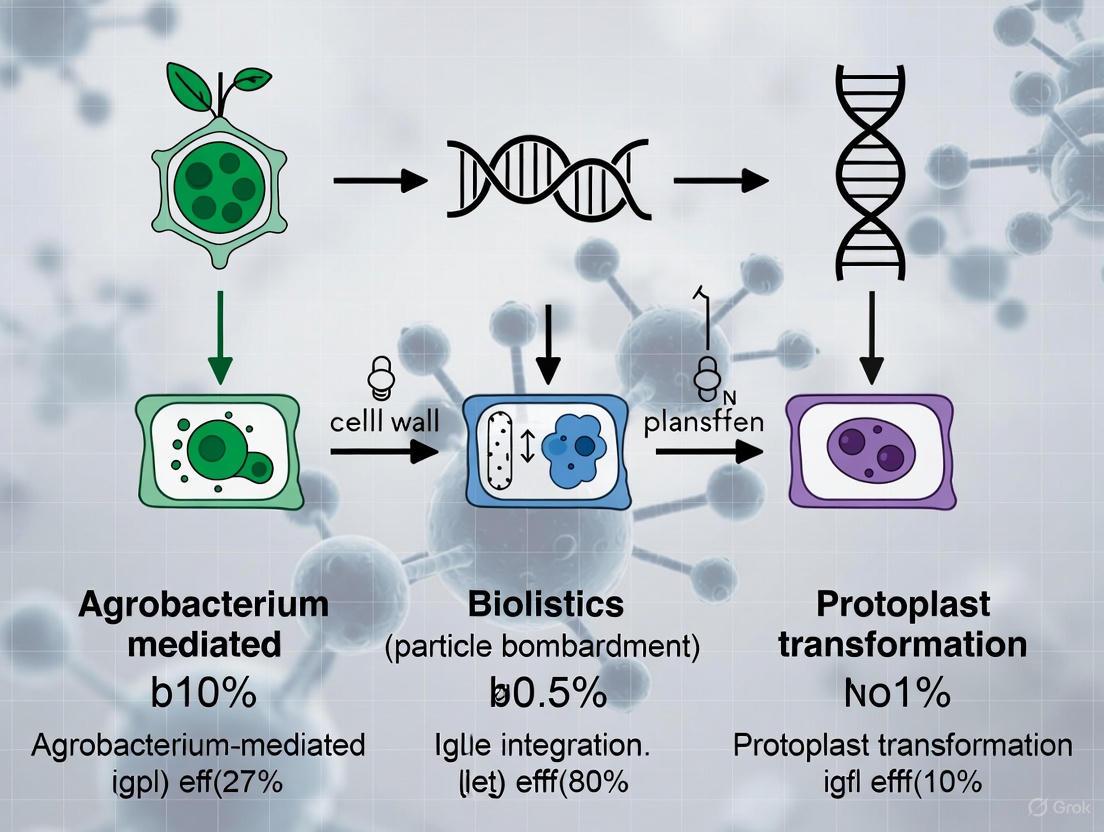
Plant genetic transformation is a foundational technique in modern plant biotechnology, enabling the introduction of foreign genes into a plant's genome to confer new traits such as disease resistance, improved nutritional content, or environmental resilience [1]. These methods are broadly categorized into two distinct groups: direct gene transfer methods and bio-mediated transformation methods [1]. Direct gene transfer techniques, such as microprojectile bombardment and protoplast methods, involve the physical or chemical delivery of DNA directly into plant cells without a biological vector. In contrast, bio-mediated transformation primarily utilizes biological agents like Agrobacterium tumefaciens or viruses to facilitate gene transfer [1]. The choice between these approaches depends on multiple factors, including the plant species, the intended application, and the available resources, with each category offering distinct advantages and limitations in terms of efficiency, cost, and technical complexity [20].
Direct Gene Transfer Methods
Core Principles and Techniques
Direct gene transfer methods are characterized by the physical or chemical delivery of foreign DNA directly into plant cells, bypassing biological vectors. A key advantage of these methods is their ability to transform a wide range of species and tissue types, making them particularly valuable for plants recalcitrant to Agrobacterium-mediated transformation [21] [20].
- Microprojectile Bombardment (Biolistics): This technique involves coating microscopic particles of gold or tungsten with DNA and propelling them into plant cells or tissues using high-pressure helium or gunpowder [21] [20]. The process can deliver various biological cargoes, including DNA, RNA, and proteins (such as CRISPR-Cas ribonucleoproteins), independent of tissue type or plant genotype [21]. A recent innovation, the Flow Guiding Barrel (FGB), has been developed to address traditional biolistic inefficiencies. This device optimizes gas and particle flow, achieving a 22-fold enhancement in transient transfection efficiency and significantly improving stable transformation frequency in maize [21].
- Protoplast-Mediated Transformation: This method involves the enzymatic removal of plant cell walls to create naked protoplasts, followed by the introduction of DNA using polyethylene glycol (PEG) or electroporation to facilitate membrane permeability [20]. While this system is highly efficient for transient gene expression studies, the regeneration of fertile plants from protoplasts remains technically challenging and species-dependent [1].
- Other Direct Methods: Additional techniques include liposome-mediated transfer, electroshock conversion, and the pollen tube pathway [1]. The pollen tube pathway, for instance, involves injecting exogenous DNA into the ovary after pollination; the pollen tube then acts as a conduit for the DNA to enter the fertilized egg, achieving transformation efficiencies of up to 2.54% in certain species like Paphiopedilum Maudiae [1].
Experimental Protocol: Microprojectile Bombardment
The following workflow details the key steps for stable plant transformation via microprojectile bombardment, optimized using the Flow Guiding Barrel (FGB) device [21].
- Microcarrier Preparation: Suspend 1µg of gold particles (0.6 µm diameter) in 10µL of sterile water. Add 1µg of plasmid DNA (e.g., pLMNC95 for GFP expression), 10µL of 2.5M CaCl₂, and 4µL of 0.1M spermidine. Vortex continuously for 3-5 minutes to coat the particles with DNA.
- Precipitation and Washing: Centrifuge the mixture briefly, remove the supernatant, and wash the DNA-gold pellets three times with 100% ethanol. Resuspend the final pellet in 20µL of 100% ethanol.
- Target Tissue Preparation: For maize transformation, isolate 100 immature embryos (1.0-1.5 mm in size) from the B104 inbred line and place them scutellum-side up on a bombardment plate containing osmotic adjustment medium.
- Bombardment Parameters: Use a PDS-1000/He system equipped with the FGB device. Set the helium pressure to 650 psi and the target distance to 6 cm. Execute a single bombardment per plate.
- Post-Bombardment Culture and Selection: Transfer bombarded embryos to callus induction medium. After a 7-day resting period, transfer the embryos to a selection medium containing herbicides or antibiotics to identify stably transformed events. Transgenic plantlets can be regenerated within 10-12 weeks.
Research Reagent Solutions for Direct Gene Transfer
The following table lists essential reagents and their specific functions in direct gene transfer protocols.
| Research Reagent | Function in Experiment |
|---|---|
| Gold microparticles (0.6 µm) | Microcarriers for DNA delivery into cells [21] |
| Spermidine | Binds DNA to microcarrier particles [21] |
| CaCl₂ (Calcium Chloride) | Co-precipitant for DNA adhesion to microcarriers [21] |
| Plasmid DNA (e.g., pLMNC95) | Vector carrying gene of interest and selectable marker [21] |
| Acetosyringone | Phenolic compound that can enhance DNA delivery in some direct methods |
Bio-mediated Transformation Methods
Core Principles and Techniques
Bio-mediated transformation harnesses the natural genetic engineering capabilities of biological vectors, primarily Agrobacterium tumefaciens, to transfer and integrate foreign DNA into the plant genome [1] [20].
- Agrobacterium tumefaciens-Mediated Transformation: This method utilizes a disarmed soil bacterium that naturally transfers a segment of its tumor-inducing (Ti) plasmid, known as T-DNA, into the plant genome [20] [22]. The process begins with the bacterium sensing plant wound signals, such as acetosyringone, which activates its virulence (vir) genes [23] [22]. The T-DNA, which can be engineered to carry genes of interest, is then excised, transported, and integrated into the plant's nuclear DNA [20]. This method is popular due to its relatively low cost, ability to transfer large DNA fragments, and tendency to produce transgenic plants with low-copy-number, clean integration events [20] [22]. Its success has been extended from dicots to many monocots, including major cereals [20].
- Virus-Mediated Transformation: While less commonly used for stable transformation, plant viruses can be engineered as vectors for transient gene expression and functional studies, though they are not a primary focus of this comparison guide [1].
- Agrobacterium rhizogenes-Mediated Transformation (Hairy Root System): This variation uses a related bacterium that transfers root-inducing (Ri) plasmid DNA, leading to the development of transgenic "hairy roots" [23] [24]. This system is highly valuable for functional genomics studies, particularly for investigating root biology and producing secondary metabolites. A recent study in Liriodendron hybrid demonstrated its efficacy, achieving transformation efficiencies of 15.51% to 60.63% across different genotypes using the K599 strain [24].
Experimental Protocol: Agrobacterium-Mediated Transformation
This protocol outlines the highly efficient transformation of photosynthetic Arabidopsis suspension cells, achieving nearly 100% transient infection rates [22].
- Agrobacterium Preparation: Inoculate the hypervirulent A. tumefaciens strain AGL1 (harboring the binary vector with the gene of interest, e.g., GFP) from a glycerol stock into YEB medium with appropriate antibiotics. Grow the main culture in AB-MES medium supplemented with 200 µM acetosyringone at 28°C until OD₆₀₀ reaches 0.3-0.5. Harvest bacterial cells by centrifugation and resuspend in ABM-MS medium to an OD₆₀₀ of 0.8 [22].
- Plant Material Preparation: Subculture photosynthetically active green Arabidopsis suspension cells in MS1 medium for 4-5 days to reach the mid-exponential growth phase [22].
- Co-cultivation: Wash the plant suspension cells twice with ABM-MS medium and adjust the packed cell volume to 70%. Mix the washed plant cells with the prepared Agrobacterium suspension and 200 µM acetosyringone. Plate the mixture onto solid ABM-MS medium containing 8 g/L plant agar and 0.05% (w/v) Pluronic F68. After drying, seal the plates and incubate at 24°C under continuous light for 2 days [22].
- Elimination of Agrobacterium and Regeneration: Following co-cultivation, wash the plant cells thoroughly with ABM-MS medium containing 250 µg/mL ticarcillin to eliminate residual Agrobacterium. For stable transformation, transfer the cells to a regeneration medium with a selection agent (e.g., kanamycin) to select for transformed events [22].
Research Reagent Solutions for Bio-mediated Transformation
The following table lists essential reagents and their specific functions in bio-mediated transformation protocols.
| Research Reagent | Function in Experiment |
|---|---|
| Agrobacterium Strain (e.g., AGL1, EHA105) | Engineered bacterial vector for T-DNA delivery [22] [24] |
| Acetosyringone | Phenolic compound that induces Agrobacterium vir gene expression [22] |
| Binary Vector (e.g., pRI101, pCAMBIA) | Plasmid carrying gene of interest between T-DNA borders [24] |
| Pluronic F68 | Surfactant that enhances transformation efficiency [22] |
| Antibiotics (e.g., Kanamycin, Ticarcillin) | Selection for transformed plants and elimination of Agrobacterium [22] |
Comparative Analysis of Transformation Methods
The following table provides a structured comparison of the key performance metrics and characteristics of direct and bio-mediated transformation methods, synthesizing data from recent studies.
| Feature | Direct Gene Transfer (Biolistics) | Bio-mediated (Agrobacterium) |
|---|---|---|
| Mechanism | Physical/chemical force delivers DNA [20] | Biological T-DNA transfer from bacterium [20] |
| Primary Technique | Microprojectile Bombardment [21] | A. tumefaciens-mediated transfer [1] |
| Host Range | Very broad; genotype-independent [21] | Broad, but efficiency varies by species [20] |
| Cargo Type | DNA, RNA, Proteins (e.g., CRISPR RNP) [21] | Primarily DNA (plasmids, T-DNA) [20] |
| Typical Integration | Complex, multi-copy insertions possible [21] [23] | Cleaner, low-copy-number, defined ends [20] |
| Transformation Efficiency (Example) | 10-fold increase in stable maize transformation with FGB [21] | Up to ~100% transient rate in optimized suspension cells [22] |
| Transgene Structure | Can be fragmented or rearranged [21] | Fewer rearrangements [20] |
| Relative Cost & Skill | High (specialized equipment) [23] | Low (minimal specialized equipment) [20] |
| Key Advantage | Delivers diverse cargo to recalcitrant species [21] | High-quality integration, low cost [20] [22] |
| Key Limitation | Complex transgene integration patterns [23] | Genotype-dependent efficiency, host range limits [1] |
Workflow Diagrams of Core Methodologies
Agrobacterium-Mediated Transformation Workflow
Biolistic Transformation Workflow
The strategic selection between direct and bio-mediated transformation methods is paramount for the success of plant genetic engineering projects. As the comparative data illustrates, bio-mediated methods, particularly Agrobacterium-mediated transformation, often provide superior integration quality and are more cost-effective for a wide range of dicot and monocot species [20] [22]. Conversely, direct methods like microprojectile bombardment offer an indispensable solution for genotype-independent transformation and the delivery of non-DNA cargo, such as CRISPR-Cas9 ribonucleoproteins for DNA-free genome editing [21].
The ongoing innovation in both categories, such as the development of the Flow Guiding Barrel for biolistics [21] and hypervirulent Agrobacterium strains for high-efficiency transformation [22], continues to push the boundaries of plant biotechnology. The choice of method ultimately hinges on the specific requirements of the experiment, including the target plant species, the desired transgene structure, the type of cargo, and the available laboratory resources. A comprehensive understanding of the principles, protocols, and comparative performance of these major transformation categories provides researchers with the foundational knowledge necessary to design and execute effective genetic transformation strategies.
The Critical Link Between Regeneration Capacity and Transformation Success
Regeneration capacity—the ability of plant cells to form new tissues or organs—is a fundamental biological process and the cornerstone of successful plant genetic transformation. This capability enables plants to repair themselves after damage and serves as the cytological foundation for in vitro plant regeneration systems used in genetic engineering [1]. For researchers aiming to develop genetically modified crops, the regeneration potential of explants often represents the primary bottleneck in the transformation pipeline [1] [25].
The efficiency of plant genetic transformation is highly dependent on species, genotypes, and explant types [25] [26]. While model plants like Arabidopsis and tomato consistently achieve high transformation efficiency, essential cereals such as maize and wheat show significantly lower efficiency and often require labor-intensive methods using immature embryos as explants [25]. This review examines the critical relationship between regeneration capacity and transformation success, comparing performance across species and methods while providing experimental data and protocols to guide researcher decision-making.
The Regeneration-Transformation Nexus: Key Mechanisms
Plant regeneration in vitro typically follows a biphasic process: (1) acquisition of cell pluripotency on auxin-enriched callus-inducing medium (CIM), and (2) de novo shoot or root regeneration on cytokinin-enriched shoot-inducing medium (SIM) [27]. Successful transformation depends on optimizing both phases, which are regulated by complex interactions between phytohormones, transcription factors, and signaling pathways [27] [25].
Molecular Regulators of Regeneration Capacity
Several key molecular pathways have been identified as critical regulators of plant regeneration capacity:
1. WOX Transcription Factors WUSCHEL-related homeobox (WOX) transcription factors control stem cell fate in meristems [25]. WOX5 is a key regulator of pluripotency acquisition in callus formation [25]. In wheat, TaWOX5 expression dramatically improved transformation efficiency, achieving up to 94.5% in readily transformable varieties and increasing efficiency from 5.8% to 55.4% in the recalcitrant variety Jimai22 [25].
2. BBM-WUS Morphogenic Factors The BABY BOOM (BBM) and WUSCHEL (WUS) transcription factors activate cell proliferation and morphogenesis during somatic embryogenesis [25]. In maize, ZmBBM expression increases callus transformation efficiency, while ZmWUS2 stimulates somatic embryo formation [25]. Their combination further enhances transformation efficiency, enabling direct Agrobacterium-mediated transformation of mature seed-derived embryo axes while eliminating dependence on immature embryo quality [25].
3. Small Signaling Peptides Recent research has identified small signaling peptides as novel regulators of regeneration capacity [27]. CLAVATA3/EMBRYO SURROUNDING REGION-RELATED (CLE) peptides negatively regulate shoot regeneration through CLAVATA1 (CLV1) and BARELY ANY MERISTEM1 (BAM1) receptors [27]. Conversely, REGENERATION FACTOR1 (REF1) peptide promotes regeneration by binding to PEPR1/2 ORTHOLOG RECEPTOR-LIKE KINASE 1 (PORK1) and activating WOUND-INDUCED DEDIFFERENTIATION 1 (WIND1) expression [27].
The following diagram illustrates the core regulatory pathways governing plant regeneration capacity:
Comparative Analysis of Regeneration and Transformation Efficiency
Genotype-Dependent Regeneration Capacity
Substantial evidence indicates that regeneration capacity and transformation efficiency vary significantly across species and genotypes, with many elite varieties being particularly recalcitrant to genetic transformation [25]. The following table summarizes comparative regeneration and transformation efficiencies across diverse plant species:
Table 1: Comparative Regeneration and Transformation Efficiencies Across Plant Species
| Species/Genotype | Explant Type | Regeneration Efficiency | Transformation Efficiency | Key Factors |
|---|---|---|---|---|
| Wheat (Triticum aestivum) | ||||
| Fielder (model) | Immature embryos | High | Up to 45.3% [25] | Genotype, explant quality |
| Jimai22 (elite) | Immature embryos | Low | 2.7-5.8% [25] | High recalcitrance |
| Jimai22 + TaWOX5 | Immature embryos | Enhanced | 55.4% [25] | Morphogenic factor |
| Soybean (Glycine max) | ||||
| Model genotypes | Various | High | Transformable [25] | Genotype flexibility |
| Elite varieties | Various | Low | Rarely successful [25] | High recalcitrance |
| Grapevine (Vitis spp.) | ||||
| Thompson Seedless | Meristematic bulk | ~100% [26] | High [26] | High competence |
| Kober 5BB | Meristematic bulk | ~100% [26] | Callus only [26] | Limited regeneration |
| 110 Richter | Meristematic bulk | Lower shoots/explant [26] | Callus only [26] | Limited regeneration |
| Oil Palm (Elaeis guineensis) | Embryogenic calli | Variable | 0.7-1.5% [28] | Monocot recalcitrance |
Transformation Method Efficiency Comparison
Different genetic transformation methods yield varying efficiencies based on plant species and regeneration capacity:
Table 2: Transformation Method Efficiency Comparison
| Transformation Method | Key Features | Ideal For | Limitations | Reported Efficiency |
|---|---|---|---|---|
| Agrobacterium-mediated | Biological vector, T-DNA integration, relatively simple [1] | Dicots, some monocots, model species [1] [28] | Host range limitations, genotype dependence [1] | Varies widely: 0.7-94.5% based on species and genotype [25] [28] |
| Pollen-tube Pathway | In planta transformation, avoids tissue culture [1] | Species with accessible pollen tubes (cotton, melon) [1] | Limited to specific developmental stages, low efficiency [1] | ~2.54% (Paphiopedilum) [1] |
| Ternary Vector Systems | Additional virulence genes, immune suppressors [29] | Recalcitrant crops (maize, sorghum, soybean) [29] | More complex vector design | 1.5-21.5× improvement over standard methods [29] |
| Morphogenic Factor-enhanced | BBM, WUS, WOX genes promote regeneration [25] | Recalcitrant genotypes, elite varieties [25] | Potential pleiotropic effects, requires transgene excision [25] | Up to 94.5% in wheat with TaWOX5 [25] |
Experimental Protocols for Assessing Regeneration-Transformation Competence
Standardized Agrobacterium-Mediated Transformation Protocol
The following detailed methodology has been successfully applied for tomato and grapevine transformation, with modifications possible for other species:
1. Explant Preparation and Meristematic Bulk Induction
- Source Material: Collect shoot tips from sterile in vitro-grown plants [26].
- Culture Conditions: Place explants on media containing increasing concentrations of cytokinins to induce meristematic bulk (MB) formation [26].
- Duration: 3-9 weeks with regular subculturing to maintain meristematic state [26].
2. Agrobacterium Preparation and Inoculation
- Strain Selection: EHA105 is commonly employed for grapevine transformations [26]. Other strains (e.g., LBA4404) may be species-dependent.
- Vector System: Binary vectors (e.g., pK7WG2) carrying selectable markers (nptII for kanamycin resistance) and reporter genes (eGFP) [26].
- Inoculation: Prepare Agrobacterium suspension to OD₆₀₀ of 0.5-1.0 in liquid medium [30]. Immerse MB slices for 15-30 minutes [26].
3. Co-cultivation and Selection
- Co-cultivation Period: 2-3 days on appropriate medium in dark conditions [30] [26].
- Selection Medium: Transfer explants to shoot induction medium containing appropriate antibiotics (e.g., 70 mg/L kanamycin for grapevine) [26] and bacteriostat (e.g., cefotaxime) to eliminate Agrobacterium [30].
- Visual Screening: Enhanced GFP fluorescence can be used alone or combined with antibiotic selection for identifying transformed tissues [26].
4. Regeneration and Plant Recovery
- Shoot Induction: Maintain selected tissues on shoot induction medium with regular subculturing every 2-3 weeks [30].
- Rooting: Transfer developed shoots to rooting medium, typically containing auxins like IAA or NAA [30].
- Acclimatization: Transfer rooted plantlets to soil under high-humidity conditions before moving to standard growth environments [30].
The experimental workflow for assessing regeneration and transformation competence is illustrated below:
Enhancement Strategies for Recalcitrant Genotypes
For genotypes with inherently low regeneration capacity, several enhancement strategies have proven effective:
1. Morphogenic Gene Overexpression
- Introduce genes such as TaWOX5, BBM, or WUS under regulated promoters [25].
- Use tissue-specific or inducible promoters to avoid pleiotropic effects [25].
- Implement "altruistic" transformation systems where morphogenic genes are transiently expressed in neighboring cells to stimulate somatic embryogenesis [25].
2. Small Signaling Peptide Application
- Apply synthetic REF1 peptide to enhance callus formation and shoot regeneration in tomato, soybean, wheat, and maize [27].
- Optimize concentration for dose-responsive enhancement (typically 0.1-10 μM) [27].
3. Ternary Vector Systems
- Employ advanced vector systems with accessory virulence genes and immune suppressors [29].
- These systems overcome intrinsic transformation barriers in recalcitrant crops [29].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Essential Research Reagents for Regeneration and Transformation Studies
| Reagent Category | Specific Examples | Function/Application | Key Considerations |
|---|---|---|---|
| Agrobacterium Strains | EHA105, LBA4404, GV3101 [30] [26] | T-DNA delivery, host range specificity | Strain selection affects transformation efficiency; EHA105 shows broad efficacy [26] |
| Vector Systems | Binary vectors (pK7WG2), Ternary vectors [26] [29] | Gene delivery, selection, expression | Ternary vectors with virulence genes enhance efficiency in recalcitrant species [29] |
| Selection Agents | Kanamycin (50-100 mg/L), Hygromycin [26] | Selective pressure for transformed tissues | Concentration must be optimized per species/genotype to minimize escapes [26] |
| Visual Reporters | eGFP, GUS [26] | Non-destructive screening of transformed tissues | Enables early identification, reduces antibiotic dependence [26] |
| Morphogenic Regulators | TaWOX5, BBM, WUS, GRF-GIF [25] | Enhance regeneration competence, overcome genotype limitations | May cause pleiotropic effects; use inducible systems recommended [25] |
| Signaling Peptides | REF1, CLE peptides, RALF33 [27] | Modulate regeneration pathways, enhance transformation | Dose-responsive effects; REF1 shows cross-species efficacy [27] |
| Hormone Stocks | Auxins (2,4-D, IAA), Cytokinins (BAP, TDZ) [27] [25] | Regulate callus formation and shoot regeneration | Balance and timing critical for phase transition in regeneration [27] |
Regeneration capacity stands as the critical determinant of plant genetic transformation success. The comparative data presented demonstrates substantial variability in both regeneration competence and transformation efficiency across species and genotypes. While technical aspects of gene transfer have advanced significantly, the fundamental biological constraint remains the innate ability of plant cells to regenerate into whole organisms.
Emerging strategies focusing on morphogenic regulators and signaling pathways offer promising avenues for overcoming regeneration limitations. The targeted manipulation of WOX transcription factors, BBM-WUS combinations, and small signaling peptides like REF1 has already demonstrated remarkable success in enhancing transformation efficiency, particularly in recalcitrant species and elite cultivars.
For researchers pursuing plant genetic engineering, particularly with applications in crop improvement, prioritizing regeneration capacity assessment in early experimental design is essential. The protocols and reagents detailed in this review provide a foundation for systematically evaluating and enhancing this critical trait, ultimately enabling more efficient genetic modification across diverse plant species.
A fundamental challenge constrains advances in plant biology and crop improvement: many elite crop varieties are recalcitrant to genetic transformation [25] [1]. This genotype dependency creates a significant bottleneck, as the most commercially valuable cultivars often prove the most difficult to modify genetically, while transformation protocols work efficiently only in a few, often non-elite, "model" genotypes [25] [31]. For instance, in wheat, transformation efficiency can range from a mere 2.7% in the commercial variety Jimai22 to 45.3% in the model genotype Fielder, with some agriculturally important varieties like Aikang58 and Jing411 failing to produce transgenic plants entirely [25] [17]. Similarly, in soybean, model genotypes are readily transformed, whereas elite commercial varieties such as Heihe43 and Zhonghuang13 are rarely transformed successfully [25]. This article examines the comparative efficiency of different approaches to overcoming this recalcitrance, focusing on experimental data and underlying molecular mechanisms.
Quantitative Comparison of Transformation Efficiency Across Methods and Genotypes
The extent of genotype dependency is starkly visible in comparative transformation studies. The following table summarizes documented transformation efficiencies for various crops, highlighting the disparity between model and elite varieties.
Table 1: Documented Transformation Efficiencies in Different Plant Species and Genotypes
| Plant Species | Genotype/Variety | Transformation Method | Reported Efficiency | Key Factor Influencing Efficiency | Citation |
|---|---|---|---|---|---|
| Wheat (Triticum aestivum) | Fielder (Model) | Agrobacterium-mediated | 45.3% | Baseline genotype susceptibility | [25] |
| Jimai22 (Elite) | Agrobacterium-mediated | 2.7% - 5.8% | Baseline genotype recalcitrance | [25] | |
| Jimai22 (Elite) | Agrobacterium-mediated + TaWOX5 | 55.4% | Morphogenic factor (WOX) | [25] [17] | |
| Multiple Elites (Baj, Kachu, etc.) | Particle Bombardment + GRF4-GIF1 | 5% - 13% | Morphogenic factor (GRF-GIF) | [32] | |
| Barley (Hordeum vulgare) | GanPi 6 & L07 (Compatible) | Agrobacterium-mediated (MDEC) | 53.2% - 56.2% | Compatible genotype | [33] |
| Hong 99 (Recalcitrant) | Agrobacterium-mediated (MDEC) | 5.4% | Recalcitrant genotype | [33] | |
| Common Bean (Phaseolus vulgaris) | Various | Agrobacterium-mediated | 0.5% - 28.6% | Protocol optimization (e.g., sonication, vacuum, hormones) | [31] |
| Maize (Zea mays) | Recalcitrant Genotypes | Agrobacterium-mediated + BBM-WUS | Significant increase reported | Morphogenic factor (BBM-WUS) | [25] [17] |
The data demonstrates that leveraging developmental regulators like WOX, BBM-WUS, and GRF-GIF can dramatically enhance efficiency in otherwise recalcitrant backgrounds, in some cases raising it to levels comparable with model genotypes [25] [32] [17].
Molecular Mechanisms of Recalcitrance and Susceptibility
The genotype-dependent response to transformation is governed by complex molecular pathways. Research has identified several key mechanisms.
The Role of Developmental (Morphogenic) Factors
A primary pathway to recalcitrance is the inability of explant cells to dedifferentiate and regenerate. Key morphogenic transcription factors that masterfully regulate this process have been identified and utilized to overcome this barrier [25] [17].
- WOX Transcription Factors: WUSCHEL-related homeobox (WOX) proteins are crucial for establishing cellular pluripotency. In wheat, overexpression of TaWOX5 increased transformation efficiency in the recalcitrant variety Jimai22 from 5.8% to 55.4% [25] [17]. It is believed that WOX factors modulate auxin and cytokinin responsiveness, which are critical for callus formation and shoot regeneration [25].
- BBM and WUS: The combination of the transcription factors BABY BOOM (BBM) and WUSCHEL (WUS) has been highly effective in monocots. In maize, their co-expression stimulates somatic embryogenesis, enhancing transformation efficiency and even enabling the transformation of mature seed-derived tissues, bypassing the need for immature embryos [25] [17]. A key challenge is that constitutive expression can cause developmental abnormalities, leading to strategies like the "altruistic" transformation system where transient WUS2 expression in some cells stimulates embryogenesis in neighboring, transformed cells [25].
- GRF-GIF Complexes: GROWTH-REGULATING FACTORS (GRFs) interacting with GRF-INTERACTING FACTORS (GIFs) are potent promoters of cell proliferation. A fusion protein of TaGRF4-TaGIF1 was used to achieve a 60-fold increase in transformation frequency in elite bread wheat cultivars, enabling efficient genome editing in previously recalcitrant lines [32].
Diagram 1: Simplified signaling pathway of morphogenic factors in transformation
Innate Plant Defense and Susceptibility Pathways
Beyond regeneration capacity, a plant's innate cellular environment determines its susceptibility to Agrobacterium infection. Transcriptomic studies comparing compatible and recalcitrant barley genotypes during infection revealed significant expressional variation in genes involved in pyruvate metabolism, plant hormone signal transduction, and DNA replication [33]. Furthermore, specific genes have been identified that act as global regulators of susceptibility.
- The MTF1 Pathway: A key discovery identified the Myb Transcription Factor 1 (MTF1) in Arabidopsis as a suppressor of transformation susceptibility [34]. Agrobacterium-secreted cytokinins trigger a signaling cascade that represses MTF1, which in turn leads to the increased expression of AT14A, a protein that may facilitate bacterial attachment to the plant cell [34]. Mutants with suppressed MTF1 are hyper-susceptible to transformation, highlighting this pathway's central role in genotypic recalcitrance.
Diagram 2: The MTF1 susceptibility pathway in Agrobacterium transformation
Experimental Protocols for Overcoming Recalcitrance
Protocol 1: Exploiting Morphogenic Factors for Wheat Transformation
A study successfully edited genes in elite bread wheat cultivars by incorporating the GRF4-GIF1 chimera [32].
- Key Experimental Workflow:
- Plant Material: Immature embryos were harvested from elite wheat cultivars (Baj, Kachu, Morocco, etc.) and the model cultivar Fielder.
- Vector Design: A binary vector containing a TaGRF4-TaGIF1 fusion gene driven by a maize ubiquitin promoter was used.
- Transformation: Immature embryos were transformed with the vector using the particle bombardment method.
- Regeneration: Transformed calli were regenerated on standard media. The presence of the GRF4-GIF1 protein dramatically improved regeneration efficiency.
- Genome Editing: Simultaneously, constructs containing CRISPR-Cas9 and guide RNAs targeting genes for leaf rust (Lr67) and powdery mildew (TaMLO) resistance were co-delivered.
- Result: Transformation frequency increased nearly 60-fold with the GRF4-GIF1-containing vectors compared to the control, ranging from ~5% to 13% across elite cultivars. Gene editing efficiency was high, with all three homeologs of the target genes successfully knocked out [32].
Diagram 3: Experimental workflow for GRF-GIF enhanced wheat transformation
Protocol 2: In planta Transformation to Bypass Tissue Culture
In planta methods represent a fundamentally different approach, aiming to transform plants with no or minimal tissue culture steps, thereby avoiding the regeneration bottleneck altogether [19] [35].
- Key Experimental Workflows:
- Floral Dip: Whole plants, typically at an early flowering stage, are submerged in a solution containing Agrobacterium and a surfactant. The bacteria transform the developing female gametophytes, leading to transgenic seeds in the next generation [19]. This method is famously successful in Arabidopsis.
- Pollen Transformation: Gene-editing tools are delivered into pollen grains via methods like electroporation or Agrobacterium infiltration. The transformed pollen is then used for pollination to produce edited seeds [35].
- Meristem Transformation: The shoot apical meristem (SAM) of embryos, seedlings, or mature plants is targeted by Agrobacterium or particle bombardment. The transformed meristematic cells can give rise to whole edited plants or germ cells [19] [35].
- Result: These methods are often more genotype-independent and avoid somaclonal variation. While efficiency can be variable and optimization is required for new species, they offer a simpler, faster, and more accessible alternative to traditional methods, particularly promising for perennial and recalcitrant crops [19] [35].
The Scientist's Toolkit: Key Research Reagents and Solutions
The following table catalogs essential reagents and materials identified in the search results as critical for tackling genotype recalcitrance.
Table 2: Key Research Reagent Solutions for Overcoming Transformation Recalcitrance
| Reagent / Material | Type | Primary Function in Research | Example Use Case |
|---|---|---|---|
| TaWOX5 Expression Vector | Morphogenic Gene Construct | Enhances cellular pluripotency and callus formation; reduces genotype dependency. | Increased wheat transformation in Jimai22 from 5.8% to 55.4% [25] [17]. |
| ZmBBM/ZmWUS2 Expression Vectors | Morphogenic Gene Construct | Promotes somatic embryogenesis; enables transformation of mature tissues. | Enhanced transformation in maize, sorghum, and wheat; used in "altruistic" systems [25]. |
| GRF4-GIF1 Fusion Protein Vector | Morphogenic Gene Construct | Boosts plant regeneration efficiency from transformed tissues. | Achieved 60-fold increase in transformation frequency in elite wheat [32]. |
| Agrobacterium tumefaciens LBA4404 | Bacterial Strain | Standard workhorse for Agrobacterium-mediated gene delivery. | Used in compatibility studies with barley microspore-derived embryogenic calli [33]. |
| Microspore-Derived Embryogenic Calli (MDEC) | Plant Explant | Provides a uniform, synchronized, and transformable cell population. | Served as subject for comparative transcriptome analysis of genotypic response in barley [33]. |
| Cytokinin (e.g., Kinetin) | Plant Hormone | Suppresses MTF1 expression to increase plant susceptibility to Agrobacterium infection. | Potential additive to transformation protocols to broaden host range [34]. |
A Practical Guide to Transformation Techniques: From Classic to Cutting-Edge
Agrobacterium tumefaciens and related Agrobacterium species are soil-borne pathogens that have been harnessed as one of the most powerful tools in plant genetic engineering. Since the initial reports in the early 1980s using Agrobacterium to generate transgenic plants, scientists have progressively improved this "natural genetic engineer" for biotechnology purposes [36]. The unique ability of Agrobacterium to transfer DNA to plant cells has been utilized for efficient delivery of genes of interest into plant genomes, revolutionizing functional genomics studies and crop improvement programs [37]. Today, many agronomically and horticulturally important species are routinely transformed using this bacterium, with an increasing number of transgenic varieties generated by Agrobacterium-mediated as opposed to particle bombardment-mediated transformation [36].
The transformation process is highly complex and evolved, involving genetic determinants of both the bacterium and the host plant cell [36]. Originally, Agrobacterium was considered only capable of transforming dicotyledonous plants, as they are the natural hosts for the bacterium. However, this paradigm shifted in 1987 when researchers demonstrated that Agrobacterium T-DNA could be incorporated into the genome of asparagus, a monocotyledon plant [28]. This breakthrough opened possibilities for transforming economically important cereal crops, including rice, maize, wheat, and barley [36] [28]. The continuous refinement of Agrobacterium-mediated transformation protocols has solidified its position as a versatile tool for plant genetic engineering nearly half a century after its discovery [38].
Molecular Mechanisms of Transformation
Bacterial Components and T-DNA Transfer
The molecular basis of genetic transformation of plant cells by Agrobacterium involves the transfer and integration of a specific DNA region from the bacterium into the plant nuclear genome. This process is mediated by two key genetic elements: the Tumor-inducing (Ti) plasmid or rhizogenic (Ri) plasmid and bacterial chromosomal genes [36] [39]. Ti plasmids are large, ranging from 200 to 800 kbp in size, and contain several functionally critical regions [36].
The T-DNA (transferred DNA) region, approximately 10-30 kbp in native plasmids, is defined by 25-bp direct repeat border sequences that exhibit high homology [36]. These border sequences serve as recognition sites for the VirD1/VirD2 endonuclease complex that processes T-DNA from the Ti plasmid [36]. The right border generally demonstrates more importance in the transfer process due to polarity established by the covalent attachment of VirD2 protein and the direction of DNA transfer [36].
Alongside T-DNA, Ti plasmids carry virulence (vir) genes that encode proteins essential for T-DNA processing and transfer [39]. These vir genes are organized into operons (virA, virB, virC, virD, virE, virG, etc.) and function in concert to excise T-DNA from the plasmid and deliver it to plant cells [36] [39]. Additionally, chromosomal genes in Agrobacterium (chv genes) participate in bacterial attachment to plant cells [39]. The presence of wounded plant tissue triggers the activation of these virulence genes through sensing specific plant-derived molecules like phenolic compounds and sugars [40] [37].
Figure 1: Agrobacterium Transformation Mechanism Signaling Pathway
Genetic Engineering of Agrobacterium Systems
For plant transformation purposes, Agrobacterium strains and vector systems have been extensively engineered. Initial modifications involved "disarming" Ti plasmids by deleting the natural tumor-inducing T-DNA regions responsible for pathogenicity while retaining the vir genes essential for DNA transfer [37]. This resulted in disarmed strains such as GV3101 (pMP90) and LBA4404 [37].
The development of the T-DNA binary vector system significantly simplified cloning procedures [37]. This system separates the T-DNA containing the genes of interest from the vir genes, with each residing on separate compatible plasmids [39]. The binary vector contains the T-DNA with left and right borders, multiple cloning site for gene of interest, plant selectable marker, bacterial selection marker, and origins of replication for both Escherichia coli and Agrobacterium [39].
Recent advancements include ternary vector systems that incorporate an additional helper plasmid carrying extra copies of vir genes to enhance transformation efficiency [37]. Similarly, super-binary vectors containing additional vir genes on the T-DNA vector have been developed, particularly beneficial for monocot transformation [37]. Other improvements include the development of auxotrophic Agrobacterium strains, such as thymidine auxotrophic lines, which require thymidine supplementation and can be easily eliminated after co-cultivation, reducing overgrowth issues [37].
A groundbreaking recent study demonstrated that engineering binary vectors to increase their copy number in Agrobacterium through point mutations in the origin of replication significantly improves transformation efficiency [41]. This approach enhanced plant transformation by up to 100% and fungal transformation by up to 400%, addressing a key bottleneck in plant and fungal engineering [41].
Workflow and Methodologies
Standard Transformation Protocol
The Agrobacterium-mediated plant transformation process follows a systematic workflow that can be divided into distinct stages, each with specific requirements and optimization points. The basic steps include [40]:
- Gene Isolation and Vector Construction: Isolating genes of interest and creating functional transgenic constructs with appropriate expression promoters, codon optimization if needed, and marker genes for tracking gene expression.
- Plasmid Introduction into Agrobacterium: Inserting the T-DNA containing plasmid into Agrobacterium using various transformation methods.
- Plant-Bacterium Co-cultivation: Mixing transformed Agrobacterium with plant cells or tissues to allow T-DNA transfer into plant chromosomes.
- Selection and Regeneration: Growing transformed cells on selection media and regenerating genetically modified plants.
- Validation and Testing: Confirming transgene integration and expression through molecular analyses and evaluating trait performance.
The preparation of Agrobacterium competent cells is a critical step that influences overall transformation efficiency. While electroporation provides the highest transformation efficiency, freeze-thaw methods offer a cost-effective alternative suitable for high-throughput applications [42] [39]. Recent protocol miniaturization enables transformation in 50μl reactions using just 200ng of DNA, with efficient heat shock performed in thermal cyclers instead of water baths [42]. Transformed cells can be plated on six-well plates, simplifying storage and handling while maintaining efficiency sufficient for routine experiments (approximately 8 × 10³ CFU/μg DNA) [42].
High-Throughput and Automated Workflows
Recent advances have focused on developing semi-automated workflows to enable high-throughput experimentation. Researchers have optimized pipelines for Agrobacterium transformation that can be adapted to robotic automation using open-source platforms like Opentrons OT-2 [42]. This system allows up to 96 transformations per batch, significantly increasing throughput capacity.
For plant transformation itself, simplified and miniaturized protocols using six-well plates have been developed for model species like Marchantia polymorpha, reducing hands-on work and costs while maintaining efficiency [42]. These improvements enable testing approximately 100 constructs per month using conventional plant tissue culture facilities, dramatically accelerating design-build-test-learn cycles for plant biotechnology [42].
A key innovation in high-throughput workflows involves enhancing selection efficiency. For example, adding sucrose to selection media significantly improves the production of propagules like gemmae in Marchantia, accelerating the generation of isogenic plants [42]. The total time from genetic construct to stable transgenic plant ready for analysis has been reduced to just four weeks with these optimized protocols [42].
Figure 2: Experimental Workflow for Plant Transformation
Applications Across Species
Transformation Efficiency in Various Crop Plants
Agrobacterium-mediated transformation has been successfully applied to numerous plant species, though efficiency varies considerably based on the plant-bacterium combination, explant type, and protocol used. The table below summarizes transformation efficiencies and key experimental parameters for major crop species:
Table 1: Transformation Efficiency in Various Crop Species
| Plant Species | Transformation Efficiency | Key Explant Types | Optimal Agrobacterium Strains | Special Requirements |
|---|---|---|---|---|
| Oil Palm (Elaeis guineensis) | 0.7-1.5% [28] | Embryogenic callus | Not specified | Highly recalcitrant; requires extensive optimization |
| Maize (Zea mays) | 33.3% with ternary vectors [37] | Immature embryos | EHA105, LBA4404 | Ternary helper plasmids improve efficiency |
| Rice (Oryza sativa) | 14-26.4% [28] | Mature embryos, shoot apex | Multiple strains | Among first monocot cereals transformed |
| Sorghum (Sorghum bicolor) | 14-33% [28] | Immature embryos | Multiple strains | Improved with super-binary vectors |
| Soybean (Glycine max) | Varies by method [43] | Hairy roots, embryonic axes | Different strains for different methods | Whole plant transformation needed for certain promoter studies |
| Marchantia (Marchantia polymorpha) | High-throughput capable [42] | Gemmae, sporelings, thalli | GV3101 | 4 weeks from construct to transgenic plant |
Comparative Analysis of Transformation Methods
Different Agrobacterium-mediated transformation methods yield varying results depending on the target species and research objectives. A comparison study in soybean demonstrated that both in vitro and in vivo hairy root transformation systems showed significantly different efficiencies, with in vitro methods proving more efficient [43]. However, for studying root-specific and low-phosphorus induced genes, neither hairy root transformation system could replace whole plant transformation, as promoter expression patterns differed substantially between systems [43].
The choice of Agrobacterium strain significantly impacts transformation success. Strains like EHA101 and EHA105, derived from the hypervirulent strain A281, generally show broader host range and higher efficiency [37]. Recent development of thymidine auxotrophic versions of these popular strains (EHA101Thy-, EHA105Thy-) helps reduce bacterial overgrowth after co-cultivation, a common problem in transformation protocols [37].
Ternary vector systems that incorporate additional vir gene helpers consistently improve transformation frequencies compared to standard binary vectors. For maize transformation, the ternary helper pKL2299A, which carries the virA gene from pTiBo542 in addition to other vir gene operons, demonstrated 33.3% transformation frequency compared to 25.6% with the original pKL2299 version [37].
Research Reagent Solutions
Successful Agrobacterium-mediated transformation requires specific reagents and genetic components optimized for different plant species. The table below outlines essential materials and their functions in the transformation process:
Table 2: Essential Research Reagents for Agrobacterium-Mediated Transformation
| Reagent/Component | Function | Examples/Specifications |
|---|---|---|
| Agrobacterium Strains | DNA delivery vector | GV3101, EHA105, LBA4404, AGL-1 [40] [39] |
| Binary Vectors | Carry gene of interest in T-DNA | pBIN19, pTF101.1; with plant/bacterial markers [39] [37] |
| Helper Plasmids | Provide vir genes in trans | Ternary helpers (pKL2299A); super-binary vectors [37] |
| Selection Antibiotics | Select transformed bacteria/plants | Kanamycin, rifampicin; strain-dependent [39] |
| Plant Growth Regulators | Promote callus formation and regeneration | Auxins, cytokinins; species-specific formulations |
| Opines | Induce vir gene expression | Acetosyringone; enhances T-DNA transfer [40] |
| Thymidine Supplement | Support auxotrophic strains | Required for thymidine auxotrophic strains [37] |
Agrobacterium-mediated transformation continues to evolve as a cornerstone technology for plant genetic engineering and biotechnology. The method offers significant advantages over alternative transformation techniques, including lower transgene copy numbers, increased stability of inserted genes, and relatively straightforward implementation [40] [39]. Recent innovations in vector engineering, including copy number optimization and ternary systems, have substantially improved transformation efficiencies in both model and recalcitrant species [37] [41].
The development of high-throughput and semi-automated workflows has further enhanced the utility of Agrobacterium-mediated transformation for large-scale functional genomics studies and crop improvement programs [42]. These advances, combined with the integration of CRISPR/Cas9 genome editing technologies, position Agrobacterium as an increasingly versatile tool for both basic research and applied biotechnology [28] [38].
Despite these improvements, challenges remain in achieving genotype-independent transformation for many economically important crop species and forest trees [36]. Future efforts will likely focus on further optimizing the fundamental biological processes underlying T-DNA transfer and integration, potentially through manipulation of both bacterial and host plant genes [36] [38]. The continued refinement of Agrobacterium-mediated transformation promises to accelerate crop improvement and functional gene analysis in an expanding range of plant species.
The development of genetically modified crops with enhanced traits such as improved yield, nutritional quality, and stress resistance relies on effective methods for delivering genetic materials into plant cells. For decades, two primary approaches have dominated this field: Agrobacterium-mediated transformation and biolistic delivery (often called the "gene gun") [44] [1]. While Agrobacterium-mediated transformation is widely used for its efficiency and tendency to produce single-copy insertion events, its utility is limited by a narrow host range, inability to deliver non-DNA cargo, and regulatory concerns associated with using a disarmed plant pathogen [44] [45]. These limitations are particularly problematic for transforming recalcitrant crop species and for emerging applications that require DNA-free gene editing.
Biolistic delivery provides a powerful alternative that overcomes these constraints. This physical method can deliver virtually any biological cargo—including DNA, RNA, and proteins—into virtually any plant species or tissue type, independent of genotype [44]. This universality makes biolistics particularly valuable for plant species resistant to Agrobacterium infection and for delivering CRISPR ribonucleoprotein (RNP) complexes for DNA-free gene editing [44]. However, despite its critical role and widespread use since 1988, conventional biolistic technology has presented longstanding challenges including inefficiency, inconsistency, and tissue damage caused by high-velocity microprojectiles [44] [46]. These limitations have historically constrained its application and efficiency in both basic research and crop improvement programs.
Recent interdisciplinary research has now identified the fundamental causes of these limitations and developed an innovative solution. Through advanced engineering approaches applied to plant biotechnology, researchers have created a Flow Guiding Barrel (FGB) that systematically optimizes particle and gas flow dynamics within the gene gun [44] [46]. This review provides a comprehensive comparison of this technological advancement against conventional biolistic delivery systems, examining its principles, performance improvements across diverse applications, and implications for the future of plant genetic engineering.
Principles of Conventional Biolistic Delivery and Its Limitations
Fundamental Mechanisms and Process
Biolistic delivery operates on a fundamentally simple principle: microscopic particles (typically gold or tungsten) coated with genetic materials are accelerated to high velocities sufficient to penetrate the rigid plant cell walls and membranes. The standard process involves several key steps: First, gold or tungsten microparticles (typically 0.6-1.0 µm in diameter) are coated with the desired cargo, which may include plasmid DNA, RNA, or proteins [46] [47]. These coated particles are then loaded onto a macrocarrier and positioned within the gene gun chamber. When activated, the device uses high-pressure helium gas to propel the microprojectiles toward the target plant tissues [44]. Upon impact, some particles successfully penetrate cell walls and membranes, releasing their cargo inside living cells where it can become functional [44].
The universality of this physical delivery mechanism gives biolistics several distinct advantages over biological delivery methods. Unlike Agrobacterium-mediated transformation, biolistics is not constrained by host specificity and does not require specific receptor molecules on target cells [44]. This capability makes it particularly valuable for transforming monocotyledonous plants (such as corn, wheat, and rice) that are naturally resistant to Agrobacterium infection [1]. Additionally, biolistics enables the delivery of diverse cargo formats, including pre-assembled CRISPR-Cas ribonucleoprotein (RNP) complexes that can immediately perform gene editing without the need for DNA integration [44]. This DNA-free editing approach minimizes off-target effects and helps avoid regulatory hurdles associated with transgenic plants [44] [45].
Historical Limitations and Technical Challenges
Despite its conceptual simplicity and versatility, conventional biolistic delivery has suffered from significant technical limitations throughout its decades of use. Computational fluid dynamic simulations of the most widely used system, the Bio-Rad PDS-1000/He, have recently revealed the fundamental causes of these inefficiencies [44]. The small aperture of the internal barrel (10 mm in diameter) creates a severe flow restriction that results in substantial particle loss, with only approximately 21% of loaded particles actually reaching the target tissue [44] [46]. This represents a massive inefficiency in reagent utilization.
The conventional design also disrupts helium flow, producing an inconsistent diffusive flow pattern that reduces gas pressure and consequently decreases particle velocity [44]. This results in uneven distribution of microprojectiles across the target tissue, with a relatively small effective coverage area of only 1.77 cm² [44]. The inconsistent particle delivery necessitated complex experimental designs with multiple bombardments to achieve reproducible results, further complicating research workflows [48]. Additionally, the unpredictable penetration depth and distribution of particles often caused excessive tissue damage, reducing the viability of transformed cells and limiting regeneration efficiency [44] [48]. These collective limitations created a significant bottleneck for applications requiring high efficiency and reproducibility, particularly for stable transformation of recalcitrant species and for emerging genome editing applications.
The Flow Guiding Barrel (FGB): A Paradigm Shift in Biolistic Technology
Fundamental Innovation and Design Principles
The Flow Guiding Barrel represents a fundamental reengineering of the biolistic delivery system based on sophisticated computational fluid dynamic simulations. Researchers discovered that the inefficiencies of conventional gene guns stemmed from basic flow dynamics issues, which they likened to "shooting a bullet without a barrel" [46]. The FGB addresses these limitations through a specifically designed internal component that replaces the standard spacer rings in the Bio-Rad PDS-1000/He system [44]. Key design parameters, including the barrel's diameter and length, were systematically optimized using both simulations and empirical testing to maximize performance [44].
The FGB functions by transforming the disrupted, diffusive gas flow of conventional systems into a uniform, laminar flow pattern that efficiently directs particles toward the target [44]. This optimized flow dynamics enables nearly 100% of loaded particles to reach their intended destination, a dramatic improvement over the 21% delivery efficiency of conventional systems [46]. Additionally, the FGB generates both higher particle velocities and a four-fold larger target area (7.07 cm² compared to 1.77 cm²), ensuring more consistent coverage and deeper penetration into target tissues [44]. The device is fabricated using Fused Deposition Modeling 3D printing, allowing for rapid prototyping and customization while maintaining compatibility with existing gene gun systems [44].
Experimental Validation and Workflow
The development and validation of the FGB followed a rigorous interdisciplinary approach integrating engineering simulations with biological testing. The research team first employed computational fluid dynamic models to analyze helium and particle flows within the conventional gene gun system, identifying the critical flow barriers [44]. Based on these insights, they designed and fabricated the FGB device, then conducted systematic performance comparisons against the conventional system across multiple biological applications.
The experimental workflow for evaluating FGB performance encompassed both physical measurements and biological assays. To physically characterize particle delivery, researchers bombarded FITC-labeled gold particles into agarose gels and measured penetration depth and distribution patterns [44]. Biological validation included transient transformation assays using GFP-DNA constructs in onion epidermis, protein delivery studies with FITC-labeled BSA, CRISPR-Cas9 RNP editing of the F3'H gene in onion cells, viral infection efficiency tests using infectious clones of soybean mosaic virus and sugarcane mosaic virus in soybean and maize seedlings, stable transformation frequency assessments in maize B104 embryos, and in planta genome editing efficiency measurements in wheat meristems using CRISPR-Cas12a [44]. This comprehensive approach provided robust quantitative data on the FGB's performance improvements across diverse applications and plant species.
The diagram below illustrates the fundamental differences between the conventional system and the FGB-enhanced system:
Comparative Performance Analysis: FGB vs. Conventional Biolistic Delivery
Quantitative Performance Metrics Across Applications
The FGB technology demonstrates substantial improvements across diverse biological applications, as systematically quantified in multiple studies. The table below summarizes key performance metrics comparing the FGB-enhanced system to conventional biolistic delivery:
Table 1: Comprehensive Performance Comparison of FGB vs. Conventional Biolistic Delivery
| Application | Plant Material | Cargo Type | Conventional System | FGB-Enhanced System | Improvement Factor |
|---|---|---|---|---|---|
| Transient Transfection | Onion epidermis | GFP-DNA (22 ng) | 153 fluorescent cells | 3,351 fluorescent cells | 22-fold [44] |
| Transient Transfection | Onion epidermis | GFP-DNA (2.2 ng) | 153 fluorescent cells (with 22 ng) | 1,031 fluorescent cells | 7-fold (with reduced DNA) [44] |
| Protein Delivery | Onion epidermis | FITC-BSA | Baseline | 4-fold increase | 4-fold [44] |
| CRISPR-Cas9 Editing | Onion epidermis | Cas9-RNP | Baseline editing | 4.5-fold increase | 4.5-fold [44] |
| Viral Infection | Maize seedlings | SCMV infectious clone | 5% infection rate | 83.5% infection rate | 17-fold [44] [47] |
| Viral Infection | Soybean seedlings | SMV infectious clone | 66% infection rate | 100% infection rate | 1.5-fold [44] |
| Stable Transformation | Maize B104 embryos | Plasmid DNA | Baseline frequency | >10-fold increase | >10-fold [44] |
| CRISPR-Cas12a Editing | Wheat meristems (T0) | Cas12a RNP | Baseline editing | 2-fold increase | 2-fold [44] |
| CRISPR-Cas12a Editing | Wheat meristems (T1) | Cas12a RNP | Baseline editing | 2-fold increase | 2-fold [44] |
| Throughput | Maize B104 embryos | Plasmid DNA | 30-40 embryos per bombardment | 100 embryos per bombardment | 2.5-3.3 fold [44] |
Analysis of Performance Improvements Across Cargo Types
The performance data reveal that the FGB enhances delivery efficiency across all major cargo types used in plant genetic engineering. For DNA-based transformation, the most dramatic improvements are observed in transient expression assays, where the 22-fold increase in GFP-expressing cells demonstrates substantially improved delivery efficiency [44]. Importantly, this enhancement persists even with reduced DNA quantities, suggesting potential applications where minimizing transgene copy number is desirable [44]. For stable transformation, the greater than 10-fold improvement in maize B104 embryos is particularly significant for crop improvement programs, as this directly translates to increased throughput and reduced costs in generating transgenic lines [44].
For protein delivery, the 4-fold enhancement in FITC-BSA internalization confirms that the FGB improves delivery of macromolecular complexes beyond nucleic acids [44]. This capability is especially valuable for CRISPR ribonucleoprotein (RNP) delivery, where the 4.5-fold increase in editing efficiency in onion epidermal cells highlights the FGB's advantage for DNA-free genome editing applications [44]. The successful enhancement of viral infectious clone delivery (17-fold improvement in maize) further demonstrates the technology's broad utility for plant-virus interaction studies and viral vector applications [44]. Notably, the doubled editing efficiency in both T0 and T1 generations of wheat using CRISPR-Cas12a RNPs demonstrates that the FGB enables improved germline editing in polyploid species, effectively addressing the previously reported low editing efficiency problem in wheat [44] [47].
Detailed Experimental Protocols and Methodologies
FGB Implementation and Bombardment Parameters
Implementing the FGB technology requires specific modifications to standard biolistic protocols. The Flow Guiding Barrel is a 3D-printed device that seamlessly integrates with the existing Bio-Rad PDS-1000/He gene gun by replacing the internal spacer rings [44]. Key operational parameters differ significantly from conventional protocols:
- Target distance: The FGB performs optimally at longer target distances (9-12 cm) compared to conventional systems (typically 6 cm for onion epidermis) [44] [48].
- Helium pressure: Effective transformation is achieved with reduced pressures (650 psi rupture disks) compared to conventional protocols (1100 psi for onion epidermis) [48].
- Particle loading: The FGB's efficient particle guidance allows reduced gold quantities per shot while maintaining high transformation efficiency [44].
- Tissue preparation: Standard tissue preparation methods are used, but the increased efficiency enables higher throughput, with maize embryo capacity increasing from 30-40 to 100 embryos per bombardment [44].
For DNA delivery, the standard protocol involves precipitating DNA onto gold microparticles (0.6-1.0 µm diameter) using calcium chloride and spermidine as precipitating agents [44] [48]. The particle suspension is then loaded onto macrocarriers and dried under vacuum before bombardment. For RNP delivery, pre-assembled Cas protein-gRNA complexes are coated onto gold particles following similar precipitation methods [44].
Assessment Methodologies for Transformation Efficiency
Quantifying transformation efficiency across different applications requires specific assessment methodologies:
- Transient transformation assays: For GFP expression studies, tissues are examined 24-48 hours post-bombardment using fluorescence microscopy. Automated cell counting platforms like CellProfiler have been customized specifically for plant cells to improve counting accuracy and throughput [48].
- Protein delivery validation: FITC-labeled proteins are visualized microscopically, with internalization confirmed through comparison with controls [44].
- Genome editing efficiency: Editing rates are quantified using next-generation sequencing of target regions, typically 2-4 days post-bombardment for transient assays or in regenerated plants for stable editing [44].
- Stable transformation frequency: For stable transformation, tissues are transferred to selection media 1-2 days post-bombardment, with transformation frequency calculated as the number of resistant events per bombarded explant [44].
- Viral infection efficiency: For viral infectious clones, infection rates are determined by monitoring reporter gene expression (e.g., GFP) or symptom development over 1-2 weeks post-inoculation [44].
The experimental workflow below illustrates the key steps in implementing and evaluating FGB-enhanced biolistic delivery:
The Scientist's Toolkit: Essential Reagents and Materials for FGB-Enhanced Biolistics
Successful implementation of FGB-enhanced biolistic delivery requires specific reagents and materials optimized for this technology. The table below details key components and their functions based on the methodologies described in the research literature:
Table 2: Essential Research Reagents and Materials for FGB-Enhanced Biolistic Delivery
| Reagent/Material | Specification/Function | Application Notes |
|---|---|---|
| Flow Guiding Barrel | 3D-printed device; replaces internal spacer rings in Bio-Rad PDS-1000/He [44] | Compatible with existing systems; optimized dimensions critical for performance |
| Gold Microparticles | 0.6-1.0 µm diameter; inert carrier for biomolecules [44] [48] | Superior to tungsten for biological applications; various sizes for different tissue types |
| Precipitation Agents | Calcium chloride and spermidine; precipitate DNA onto gold particles [44] [48] | Standard protocol; concentration optimization may be required for specific applications |
| Helium Gas | High purity; propulsion source for microprojectiles [44] | System requires precise pressure regulation (450-1100 psi rupture disks) |
| Rupture Disks | 450-1100 psi rating; controls helium pressure [44] [48] | FGB enables effective transformation at lower pressures (650 psi) |
| GFP Reporter Plasmids | e.g., pLMNC95; visual marker for transient transformation efficiency [44] [48] | Quantitative assessment via fluorescence microscopy or automated counting |
| CRISPR Reagents | Cas proteins, guide RNAs, or pre-assembled RNPs; for DNA-free genome editing [44] | FGB significantly enhances RNP delivery efficiency (4.5-fold improvement) |
| Cell Viability Stains | Fluorescein diacetate (FDA); assesses tissue damage post-bombardment [48] | Critical for optimizing parameters to balance efficiency with tissue health |
| Automated Counting Software | Customized CellProfiler pipelines; quantifies transformed cells [48] | Specifically adapted for plant cell morphology and autofluorescence |
The development of the Flow Guiding Barrel represents a significant advancement in biolistic technology, addressing fundamental limitations that have persisted for decades. By systematically optimizing gas and particle flow dynamics through computational modeling and engineering design, the FGB enhances delivery efficiency across diverse cargo types and plant species while maintaining compatibility with existing gene gun systems [44] [46]. The documented improvements—ranging from 4-fold for protein delivery to over 20-fold for transient DNA expression—demonstrate this technology's potential to overcome critical bottlenecks in plant genetic engineering [44].
This advancement holds particular significance for emerging applications in plant genome editing. The enhanced delivery of CRISPR-Cas ribonucleoprotein complexes enables more efficient DNA-free editing, addressing regulatory concerns and facilitating the development of improved crop varieties [44] [45]. The demonstrated improvements in wheat meristem editing further suggest that FGB technology can help overcome the challenges of editing polyploid genomes, where multiple gene copies must be simultaneously modified to achieve desired traits [44] [47]. Additionally, the reduced tissue damage and increased consistency provided by the FGB's laminar flow pattern may improve regeneration efficiency from transformed tissues, particularly for recalcitrant species [44] [48].
As plant genetic engineering continues to evolve toward more precise editing techniques and broader species applicability, efficient and versatile delivery technologies like the FGB-enhanced biolistic system will play an increasingly critical role. The commercial development of this technology through startup companies ensures its accessibility to the broader research community, potentially accelerating both basic plant science and applied crop improvement efforts [46] [49]. By addressing the fundamental inefficiencies that have long constrained biolistic delivery, the Flow Guiding Barrel represents not merely an incremental improvement but a transformative advancement that expands the boundaries of what is possible in plant genetic engineering.
Plant genetic transformation is a cornerstone of modern plant biology and biotechnology, enabling functional gene studies and the development of new crop traits. However, traditional methods that rely on extensive tissue culture present significant bottlenecks due to their time-consuming nature, technical complexity, and high genotype dependence [19] [18]. In planta transformation strategies have emerged as revolutionary alternatives that minimize or eliminate tissue culture requirements, offering technical simplicity, lower costs, and greater genotype independence [19] [50].
This guide provides a comparative analysis of three prominent in planta techniques: the well-established Floral Dip method, the Pollen-Tube Pathway, and the emerging Leaf-Cutting Transformation (LCT). By objectively examining their protocols, efficiencies, and applications, we aim to equip researchers with the necessary information to select the most appropriate method for their experimental needs.
Methodologies and Experimental Protocols
Floral Dip Transformation
The floral dip method is one of the most widely used in planta techniques, particularly in Brassicaceae species [19] [51]. The standard protocol involves:
- Plant Preparation: Grow plants until the stage of early bud development and flowering.
- Agrobacterium Preparation: Culture Agrobacterium tumefaciens (e.g., strain GV3101) carrying the binary vector with the gene of interest until OD₆₀₀ reaches approximately 0.6 [51].
- Inoculum Preparation: Centrifuge bacterial culture and resuspend in infiltration medium (5% sucrose, 0.03-0.05% Silwet L-77) [51].
- Transformation: Dip developing inflorescences into the Agrobacterium suspension for 45-90 seconds with gentle agitation [51].
- Post-Treatment: Maintain dipped plants under high humidity for 24 hours, then grow to seed maturity under normal conditions.
- Selection: Harvest seeds and screen on selective medium containing appropriate antibiotics (e.g., hygromycin B) or through molecular verification [51].
Key factors influencing efficiency include Agrobacterium density, surfactant concentration, plant developmental stage, and sucrose concentration [51].
Pollen-Tube Pathway Transformation
This method exploits the natural pathway created by pollen tubes during fertilization to deliver foreign DNA into embryos [1]:
- Plant Preparation: Grow plants until flowering and perform manual pollination.
- Timing: After pollination (typically 4-24 hours, species-dependent), identify the optimal window when pollen tubes have reached the ovules but before fertilization is complete [1].
- DNA Delivery: Apply DNA solution (5-20 μL) containing the target gene to the ovary or style using microinjection or droplet application.
- Integration Mechanism: The exogenous DNA travels along the pollen tube pathway and integrates into the developing embryo genome.
- Selection: Harvest mature seeds and screen for transformants using molecular or phenotypic markers.
Transformation efficiency is highly dependent on precise timing relative to the fertilization process and the method of DNA application [1].
Leaf-Cutting Transformation (LCT)
LCT is a recently developed tissue culture-free method that utilizes the regenerative capacity of leaf tissues [52]:
- Explant Preparation: Collect healthy, mature leaves and cut into segments (e.g., 1-2 cm²).
- Agrobacterium Preparation: Culture Agrobacterium tumefaciens (e.g., strain EHA105) carrying the visual reporter Ruby system to OD₆₀₀ ≈ 0.6-0.8 [52].
- Infection: Immerse leaf segments in Agrobacterium suspension for 15-30 minutes.
- Co-cultivation: Transfer infected leaves to sterile vermiculite or similar substrate and maintain under high humidity for 2-3 days.
- Regeneration: Culture treated leaves in non-sterile conditions (e.g., vermiculite) for 8-12 weeks until bud formation occurs at the wound sites [52].
- Selection: Identify transformants using visual markers (e.g., betalain pigmentation from Ruby system) without antibiotic selection.
This method leverages the natural regenerative capacity of certain plant species, particularly those with high leaf propagation ability [52].
Table 1: Key Comparative Parameters of In Planta Transformation Methods
| Parameter | Floral Dip | Pollen-Tube Pathway | Leaf-Cutting Transformation |
|---|---|---|---|
| Target Tissue | Developing inflorescences, female gametes | Pollen tubes, fertilized ovules | Vegetative leaf tissues, meristems |
| Delivery Mechanism | Agrobacterium-mediated | Direct DNA integration | Agrobacterium-mediated |
| Typical Efficiency Range | 0.1-5% [51] | Up to 2.54% [1] | 14.2-46.65% (in perennial ryegrass) [53] |
| Regeneration Pathway | Seed development | Seed development | Direct organogenesis from leaf |
| Selection System | Antibiotic/herbicide resistance | No selection required | Visual markers (e.g., Ruby system) |
| Species Demonstrated | Arabidopsis, Brassicaceae species [51] | Cotton, melon, soybean, wheat, corn [1] | Jonquil, perennial ryegrass [53] [52] |
Comparative Efficiency Analysis
Transformation Efficiency Across Species
Transformation efficiency varies significantly among methods and across plant species, as quantified in recent studies:
Table 2: Documented Transformation Efficiencies Across Plant Species
| Plant Species | Method | Efficiency | Experimental Conditions |
|---|---|---|---|
| Descurainia sophia | Floral Dip | 1.52% | OD₆₀₀=0.6, 0.03% Silwet L-77, no acetosyringone [51] |
| Paphiopedilum Maudiae | Pollen-Tube Pathway | 2.54% | Ovary injection pre-fertilization [1] |
| Jonquil | LCT | 100% (regeneration), ~40% (transformation) | A. tumefaciens EHA105, Ruby visual marker [52] |
| Perennial ryegrass | SAAT (LCT variant) | 14.2-46.65% | Sonication-assisted, seed/meristem transformation [53] |
| Rice (mature embryos) | In Planta (seedling) | 3.5-9% | Agrobacterium-mediated, varied by cultivar [53] |
Technical Considerations and Limitations
Each method presents distinct advantages and limitations that influence their applicability:
Floral Dip advantages include technical simplicity and established protocols for Brassicaceae, while limitations include primarily effective in Brassicaceae species and sensitivity to plant developmental stage [19] [51].
Pollen-Tube Pathway advantages comprise no requirement for tissue culture and wide applicability across species, with limitations being precise timing critical and generally lower efficiency rates [1].
LCT advantages include no sterile culture requirements and utilizes readily available leaf material, while limitations are currently limited to species with high regenerative capacity and relatively long regeneration time (8-15 weeks) [53] [52].
Signaling Pathways and Molecular Mechanisms
The efficiency of in planta transformation methods is influenced by complex signaling pathways that regulate plant cell reprogramming and regeneration:
This diagram illustrates the key molecular pathways that influence regeneration efficiency across in planta methods. The WIND1, PLT, and REF1 genes activate downstream factors involved in callus formation and dedifferentiation [18] [54]. Meanwhile, WUS, BBM, and GRF-GIF complexes promote meristem formation and organ regeneration [18] [55]. These endogenous pathways can be harnessed to improve transformation efficiency across all three methods discussed.
The Scientist's Toolkit: Essential Research Reagents
Successful implementation of in planta transformation methods requires specific reagents and biological materials:
Table 3: Essential Research Reagents for In Planta Transformation
| Reagent/Material | Function | Application Examples |
|---|---|---|
| Agrobacterium tumefaciens Strains | DNA delivery vector | GV3101 (Floral Dip) [51], EHA105 (LCT) [52] |
| Silwet L-77 | Surfactant that enhances tissue penetration | Floral Dip (0.03-0.05%) [51] |
| Visual Marker Systems | Transformation confirmation without selection | Ruby system (betalain biosynthesis) for LCT [52] |
| Selective Agents | Transformed plant selection | Hygromycin B for Floral Dip [51] |
| Morphogenic Regulators | Enhance regeneration capacity | WUS2/BBM for enhancing transformation in grasses [55] |
| Sucrose Solution | Osmoticum and energy source in inoculation media | 5% sucrose in Floral Dip [51] |
| Acetosyringone | Induces vir gene expression in Agrobacterium | Varies by species and method (0-200 μM) [51] |
The comparative analysis of these three in planta transformation methods reveals a trade-off between technical simplicity, efficiency, and species applicability. Floral Dip remains the gold standard for amenable species, particularly in Brassicaceae, while the Pollen-Tube Pathway offers a tissue culture-free alternative for a broader range of species with moderate efficiency. The emerging LCT technique demonstrates remarkable potential for high-efficiency transformation in species with strong regenerative capacity, without requiring sterile conditions.
Future directions in in planta transformation will likely focus on expanding the host range of these methods through the identification and utilization of developmental regulators that enhance cellular reprogramming and regeneration capacity. The integration of genome editing technologies with simplified transformation methods will further accelerate functional genomics and precision breeding in diverse plant species [18] [50].
Virus-Induced Genome Editing (VGEd) and Agrobacterium rhizogenes-Mediated Hairy Root Systems
The advancement of plant functional genomics and molecular breeding is fundamentally reliant on efficient genetic transformation technologies. Among the various methods developed, Virus-Induced Genome Editing (VGEd) and Agrobacterium rhizogenes-mediated hairy root transformation have emerged as powerful platforms for gene function analysis and genome editing assessment. These systems offer distinct advantages for specific research applications, particularly in plant species recalcitrant to stable genetic transformation [56]. VGEd utilizes modified viral vectors to deliver genome editing components into plants, enabling targeted mutagenesis without the need for traditional tissue culture [56]. Meanwhile, the hairy root system leverages the natural ability of A. rhizogenes to transfer DNA containing root-inducing (Ri) genes into plant cells, leading to the development of transgenic roots that facilitate rapid functional gene analysis [57] [58]. This review comprehensively compares the efficiency, applications, and methodologies of these two transformative approaches, providing researchers with critical insights for selecting appropriate strategies for their plant genetic engineering endeavors.
Agrobacterium rhizogenes-Mediated Hairy Root Transformation
The hairy root transformation system utilizes the natural genetic engineering capabilities of the soil bacterium Agrobacterium rhizogenes. This pathogen transfers specific DNA segments (T-DNA) from its root-inducing (Ri) plasmid into the plant genome upon infection of wounded plant tissues [57]. The integrated T-DNA contains genes that disrupt normal plant hormone balance, leading to the prolific development of adventitious roots at the infection site [57] [59]. These transgenic roots, known as "hairy roots," exhibit rapid growth, genetic stability, and can be maintained as composite plants (non-transgenic shoots with transgenic roots) or cultured in vitro [60].
The applicability of this system has been significantly enhanced through the development of various engineered A. rhizogenes strains. Commonly used wild-type strains include ATCC15834, A4 (ATCC43057), K599 (NCPPB2659), and NCPPB1855 (also known as LBA9400) [57]. These strains have been engineered to create derivatives with improved virulence and antibiotic resistance markers, such as A4RS (resistant to rifampicin and spectinomycin) [57]. The molecular mechanism involves the transfer of T-DNA bounded by left and right border sequences into the plant cell nucleus, where it integrates into the plant genome and expresses the encoded genes, including the rol (root loci) genes essential for hairy root induction [57] [59].
Virus-Induced Genome Editing (VGEd)
Virus-Induced Genome Editing represents a revolutionary approach that combines the high efficiency of viral vectors with targeted genome editing technologies. In VGEd, plant RNA viruses are modified to serve as delivery vehicles for CRISPR/Cas components, particularly the single guide RNA (sgRNA) [56]. The system typically involves infecting transgenic plants that constitutively express Cas9 nuclease with viral vectors carrying sgRNA sequences targeting specific genomic loci [56].
The fundamental mechanism leverages the natural replication and movement capabilities of viruses within plant tissues. As the viral vector spreads systemically, it delivers the sgRNA to numerous cells, where it complexes with the Cas9 protein and induces targeted DNA double-strand breaks [56]. These breaks are subsequently repaired by the plant's endogenous DNA repair machinery, primarily through error-prone non-homologous end joining (NHEJ), resulting in targeted mutations [61]. This approach enables efficient genome editing without the need for tissue culture, making it particularly valuable for challenging plant species [56]. Commonly used viral vectors include Tobacco Rattle Virus (TRV) and Citrus Leaf Blotch Virus (CLBV), which have been successfully employed for VGEd in various plant species [56].
Comparative Efficiency Analysis
The practical utility of genetic transformation technologies is largely determined by their efficiency, which we define as the success rate in achieving the desired genetic modification. The table below provides a comprehensive comparison of key performance metrics between hairy root transformation and VIGE systems across multiple plant species, based on recent experimental studies:
Table 1: Comparative Efficiency of Hairy Root Transformation and Virus-Induced Genome Editing
| Plant Species | Technology | Transformation/Editing Efficiency | Time Required | Key Applications Demonstrated | Reference |
|---|---|---|---|---|---|
| Soybean | Hairy Root | 58-94.3% transformation frequency; 5-88.8% editing efficiency | 14-16 days | sgRNA validation, protein localization, protein-protein interaction | [59] [58] |
| Cotton | Hairy Root | 30% transformation frequency; mutations detected in 40-50% of hairy root lines | ~30 days | sgRNA validation for GhPDS gene | [60] |
| Citrullus species | Hairy Root | 90.9% of accessions showed mutations; 73.94% of roots mutated at target site | ~30 days | Validation of sgRNA efficiency for ClCIPK17 | [62] |
| Tobacco | VIGE | Successful editing of endogenous PDS gene | Not specified | Genome editing without tissue culture | [56] |
| Multiple legumes | Hairy Root | 17.7-43.3% transformation efficiency across species | 14 days | Rapid evaluation of editing efficiency | [63] |
The data reveal that hairy root transformation consistently achieves high efficiency across multiple plant species, with soybean showing particularly impressive transformation frequencies (up to 94.3%) and editing efficiencies (up to 88.8%) [59] [58]. The system's robustness is further demonstrated in Citrullus species, where 90.9% of accessions showed mutations at the target gene [62]. The time investment for hairy root transformation varies between 14-30 days, depending on the protocol and species [59] [58] [62].
While quantitative efficiency data for VIGE is more limited in the available literature, successful genome editing of endogenous genes has been demonstrated in tobacco using this approach [56]. The key advantage of VIGE lies in its ability to bypass tissue culture entirely, making it particularly valuable for species recalcitrant to transformation [56].
Experimental Protocols and Workflows
Hairy Root Transformation Protocol
The hairy root transformation system has been optimized for various plant species, with detailed protocols established for soybean and cotton that can be adapted for other species. Below is a comprehensive workflow for implementing this technology:
Diagram 1: Hairy root transformation experimental workflow
The protocol begins with plant material preparation. For soybean, seeds are surface-sterilized using chlorine gas and germinated for 1-7 days [58] [63]. Explants are typically prepared by making slant cuts in the hypocotyl or using split-imbibed seeds with the 1/3 part of cotyledon with hypocotyl excised at a 45° angle [58].
Simultaneously, A. rhizogenes strain K599 harboring the binary vector with the gene of interest is cultured in YEP or TY medium with appropriate antibiotics until it reaches an OD₆₀₀ of 0.6-1.2 [60] [58]. The bacterial cells are then pelleted by centrifugation and resuspended in co-cultivation medium, which typically contains B5 or MS salts, sucrose, plant hormones (6-BA, GA₃), and acetosyringone (40-100 μmol) to induce virulence [58] [63].
The inoculation and co-cultivation phase involves infecting the explants with the bacterial suspension for 30 minutes to several hours [59] [58]. The infected explants are then transferred to solid co-cultivation medium and incubated at 24°C under a 16-h-light/8-h-dark photoperiod for 2-5 days [58]. After co-cultivation, the explants are transferred to rooting medium containing antibiotics to suppress Agrobacterium overgrowth [58].
During the root development phase, hairy roots typically emerge within 6-14 days post-inoculation [59] [58]. Transgenic roots can be selected using visual markers (e.g., Ruby gene for red pigmentation, GFP for fluorescence) or selectable markers (e.g., herbicide resistance) [63]. Finally, molecular analyses including PCR, sequencing, and phenotypic assessments are performed on the transgenic roots to verify transformation efficiency and editing success [60] [58].
Virus-Induced Genome Editing Protocol
The VIGE protocol utilizes viral vectors to deliver genome editing components into plants. The workflow below outlines the key steps in implementing this technology:
Diagram 2: Virus-Induced Genome Editing experimental workflow
The VIGE protocol begins with viral vector construction. The sgRNA expression cassette is cloned into a modified viral vector, such as Tobacco Rattle Virus (TRV) or Citrus Leaf Blotch Virus (CLBV), ensuring that essential elements for viral replication and movement are maintained [56].
A critical requirement for conventional VIGE is the use of Cas9-overexpressing transgenic plants that constitutively express the Cas9 nuclease [56]. These plants are typically grown for 3-5 weeks until they reach an appropriate developmental stage for inoculation [56].
The viral delivery step involves inoculating the Cas9-expressing plants with the engineered viral vector. Multiple inoculation methods can be employed, including agroinfiltration, mechanical rubbing, or other species-specific inoculation techniques [56].
Following inoculation, the virus undergoes systemic spread throughout the plant, replicating in infected cells and moving through the vascular system to reach meristematic tissues [56]. During this process, the sgRNA is expressed from the viral vector and complexes with the plant-encoded Cas9 protein to form functional ribonucleoproteins.
The actual genome editing occurs when these ribonucleoproteins induce double-strand breaks at the target genomic loci, which are subsequently repaired by the plant's endogenous DNA repair mechanisms, primarily through error-prone non-homologous end joining (NHEJ) [61] [56].
Finally, editing efficiency analysis is performed by extracting DNA from newly developed tissues, amplifying the target regions by PCR, and sequencing the amplicons to detect mutations. Deep sequencing is often employed to quantify editing efficiency and characterize the spectrum of induced mutations [56].
Research Reagent Solutions
Successful implementation of either hairy root transformation or VIGE requires specific research reagents and biological materials. The table below outlines essential solutions for establishing these technologies:
Table 2: Essential Research Reagents for Hairy Root Transformation and VIGE
| Reagent Category | Specific Examples | Function and Application | Technology |
|---|---|---|---|
| Agrobacterium Strains | K599, ATCC15834, A4, A4RS | Delivery of T-DNA containing genes of interest; induction of hairy roots | Hairy Root |
| Binary Vectors | pFGC5941, pKSE401, pCAMBIA2301 | Carrying gene constructs, markers, and CRISPR components | Hairy Root |
| Visual Markers | Ruby, GFP, DsRed1 | Visual identification of transgenic roots without specialized equipment | Hairy Root |
| Selectable Markers | BAR (herbicide resistance), Kanamycin resistance | Selection of transformed tissues | Hairy Root |
| Viral Vectors | TRV, CLBV | Delivery of sgRNA in virus-induced genome editing | VIGE |
| Plant Growth Regulators | 6-BA, GA₃ | Enhance transformation efficiency during co-cultivation | Hairy Root |
| Induction Compounds | Acetosyringone (AS) | Induces Agrobacterium virulence genes | Both |
| Culture Media | B5, ½ B5, MS, ½ MS | Support plant tissue growth and transformation | Hairy Root |
The selection of appropriate A. rhizogenes strains is critical for successful hairy root transformation. Strain K599 is particularly effective for legumes including soybean and peanut, while strains ATCC15834 and A4 are broadly applicable to various plant species [57] [60] [58]. Engineered strains like A4RS offer antibiotic resistance markers that facilitate counter-selection against non-transformed bacteria [57].
Binary vectors for hairy root transformation typically contain multiple components: (1) T-DNA border sequences for DNA transfer, (2) Cas9 expression cassettes driven by constitutive promoters like CaMV 35S, (3) sgRNA expression units driven by Pol III promoters such as U6, and (4) selection or visual markers [57] [60]. Modifications such as the addition of translational enhancers like OsMac3 can significantly boost expression levels [57].
Visual markers have revolutionized transgenic root identification, with the Ruby reporter system enabling rapid, equipment-free selection of transformed roots based on intense red pigmentation [63]. Similarly, GFP and other fluorescent proteins allow visual screening under appropriate lighting [58].
For VIGE, viral vectors must be engineered to accommodate sgRNA sequences while maintaining viral replication and movement functions. TRV vectors are commonly used due to their broad host range and efficient systemic movement [56].
Culture media composition significantly impacts transformation efficiency. Research indicates that ½ B5 medium often yields superior results compared to full-strength formulations or MS-based media for hairy root induction in species like soybean [58]. The addition of acetosyringone, a phenolic compound that induces Agrobacterium virulence genes, is essential for maximizing transformation efficiency in both technologies [58] [63].
Applications in Plant Functional Genomics
Both hairy root transformation and VIGE have diverse applications in plant functional genomics and biotechnology. The table below compares their primary applications and utilities:
Table 3: Comparative Applications in Plant Functional Genomics
| Application | Hairy Root Transformation | Virus-Induced Genome Editing |
|---|---|---|
| sgRNA Validation | Excellent: Pre-screening of sgRNAs before stable transformation; 71.43-97.62% efficiency in soybean [59] | Limited: Primarily used for editing rather than validation |
| Protein Studies | Excellent: Protein subcellular localization, protein-protein interaction (BiFC), protein expression [58] | Not applicable |
| Root Biology Research | Excellent: Functional studies of root-specific genes, root-microbe interactions, nutrient uptake [59] [58] | Limited: Not specialized for root studies |
| Metabolic Engineering | Excellent: Production of valuable phytochemicals, metabolic pathway engineering [59] | Limited: Not typically used for metabolic engineering |
| High-Throughput Screening | Good: Moderate throughput for sgRNA and gene function screening [63] | Excellent: Potentially higher throughput due to viral systemic spread |
| Whole-Plant Genome Editing | Limited: Primarily generates chimeric plants with transgenic roots [59] | Excellent: Can achieve heritable edits in meristematic tissues [56] |
| Tissue Culture-Free Editing | Limited: Requires some tissue culture steps in most protocols | Excellent: Completely bypasses tissue culture requirements [56] |
Hairy root transformation excels in functional gene validation, particularly for root-specific processes. The system has been successfully employed to study root-microbe interactions, nutrient uptake, and root architecture [59] [58]. Additionally, it provides an excellent platform for protein subcellular localization and protein-protein interaction studies using techniques like bimolecular fluorescence complementation (BiFC) [58].
A particularly valuable application of hairy roots is the pre-screening of sgRNAs for CRISPR/Cas9 experiments. Research demonstrates strong correlation (Pearson correlation coefficient of 0.83) between editing efficiencies in hairy roots and stable transgenic plants, enabling researchers to identify effective sgRNAs before investing in lengthy stable transformation procedures [59]. This approach has been successfully applied in soybean, cotton, and Citrullus species [60] [59] [62].
VIGE technology offers unique advantages for whole-plant genome editing without tissue culture requirements. The systemic movement of viral vectors enables delivery of editing components to meristematic tissues, potentially generating heritable mutations [56]. This approach is particularly valuable for species recalcitrant to transformation using conventional methods.
Both technologies significantly accelerate the characterization of gene function compared to stable transformation. Hairy root systems can provide functional data within 2-4 weeks, while VIGE can potentially generate edited plants in a single generation without the need for tissue culture [58] [56].
The comparative analysis of Virus-Induced Genome Editing and Agrobacterium rhizogenes-mediated hairy root transformation reveals complementary strengths that suit different research objectives. Hairy root transformation offers a robust, well-established platform for rapid gene function analysis, particularly for root biology, protein studies, and pre-screening genome editing targets. Its high efficiency across diverse plant species, relatively simple implementation, and versatility for various applications make it invaluable for functional genomics research.
VIGE represents a transformative approach for achieving heritable genome edits without tissue culture requirements, offering particular promise for plant species recalcitrant to conventional transformation. While quantitative efficiency data is more limited for VIGE in the current literature, its potential for high-throughput editing and application in challenging species positions it as a technology with significant future impact.
Selection between these technologies should be guided by specific research goals: hairy root transformation for rapid functional analysis and sgRNA validation, and VIGE for tissue culture-free whole-plant editing. As both technologies continue to evolve, they will undoubtedly expand the frontiers of plant genetic engineering and functional genomics, enabling researchers to address increasingly complex biological questions and contribute to crop improvement efforts.
Plant genetic transformation is a cornerstone of modern biotechnology, enabling the development of crops with enhanced traits such as improved yield, disease resistance, and environmental resilience. Among the various approaches available, direct DNA transfer methods bypass biological vectors to introduce genetic material directly into plant cells. This guide provides a comparative analysis of three prominent direct delivery systems: protoplast-based transformation, electroporation, and nanoparticle-mediated techniques. Understanding the mechanisms, efficiencies, and limitations of each method is crucial for researchers selecting the optimal approach for specific applications, from basic research to commercial crop development. The continuous evolution of these technologies, particularly with advancements in genome editing tools like CRISPR/Cas9, underscores the importance of evaluating their performance parameters and integration capabilities with modern biotechnological applications [64] [65].
Direct DNA transfer methods share the common principle of introducing genetic material into plant cells without using biological intermediaries like Agrobacterium tumefaciens. These techniques physically or chemically facilitate the passage of DNA through the formidable plant cell wall and plasma membrane barriers.
Protoplast Transformation utilizes plant cells whose walls have been enzymatically removed, creating naked cells susceptible to DNA uptake. These protoplasts are typically incubated with DNA in the presence of polyethylene glycol (PEG) and calcium ions or subjected to electrical pulses. The permeable membrane allows for direct entry of DNA, leading to transient expression or stable integration upon regeneration of the cell wall and subsequent cell division [64] [66]. A significant advantage of this system is its potential for high-throughput transformation and avoidance of transgenesis when using DNA-free editing tools like ribonucleoproteins (RNPs) [66].
Electroporation involves applying a high-intensity electrical field to a suspension of plant cells or tissues. This transiently disrupts the phospholipid bilayer of the plasma membrane, creating nanoscale pores that allow macromolecules like DNA to enter the cell. While often applied to protoplasts, this method can also be adapted for intact tissues, though with lower efficiency due to the constraining presence of the cell wall [67] [68]. The process is highly variable and depends on parameters such as cell type, field strength, pulse duration, and the conductivity of the surrounding medium [67].
Nanoparticle-Mediated Delivery represents a more recent innovation in plant genetic engineering. This method employs nanoscale particles (typically <100 nm in diameter)—such as carbon-based carriers, gold nanoparticles, or liposomes—that are complexed with genetic cargo. Due to their small size and tunable surface chemistry, these nanoparticles can traverse the pores of the plant cell wall and release their cargo into the cytoplasm. This technique is particularly promising for its passive delivery mechanism, which minimizes tissue damage and demonstrates broad host-range applicability [67] [68].
Comparative Performance Analysis
The selection of an appropriate genetic transformation method depends heavily on experimental goals, target plant species, and available resources. The table below provides a detailed comparison of key performance metrics for protoplast, electroporation, and nanoparticle-mediated techniques.
Table 1: Comprehensive Comparison of Direct DNA Transfer Methods
| Performance Metric | Protoplast Transformation | Electroporation | Nanoparticle-Mediated Delivery |
|---|---|---|---|
| Core Mechanism | Chemical (PEG) or physical facilitation of DNA uptake into wall-less cells [66] | Electrical field-induced pore formation in the membrane [67] [68] | Passive diffusion and intracellular release via engineered nanoparticles [67] [68] |
| Primary Target Tissue | Protoplasts isolated from leaves, cotyledons, or callus [64] [66] [69] | Protoplasts or intact tissues with thin cell walls [67] | Diverse tissues including leaves, roots, and germinating seeds [67] [68] |
| Transformation Efficiency | High for transient expression in amenable species; stable transformation efficiency is genotype-dependent [66] | Variable; highly efficient for protoplasts, lower for walled cells [67] | Demonstrated high efficiency in model plants; rapidly improving for crops [67] [68] |
| Cargo Type & Size Limit | DNA, RNA, Proteins (RNPs); theoretically unlimited [66] | DNA, siRNA, miRNA; limited by pore size and stability [67] [68] | DNA, siRNA, dsRNA, proteins; size limited by nanoparticle loading capacity [67] |
| Tissue Damage & Cell Viability | Viability depends on gentle isolation and culture; can suffer from cell aggregation [69] | High field pulses can cause excessive cell death and overheating [67] | Generally low cytotoxicity; minimal tissue damage [67] [68] |
| Regeneration Requirement & Challenge | Required; often a major bottleneck as protocols are species-specific and can be lengthy (e.g., 15 weeks for Arabidopsis) [66] [69] | Required if using protoplasts; not always needed for transient expression in tissues [68] | Not always required; can lead to heritable edits without regeneration in some systems [68] |
| Key Limitation | Protoplast regeneration is inefficient for many species; genotype-dependent [66] | Requires standardized protoplast preparation; optimization is complex [67] | Lack of universal protocols; long-term fate of nanoparticles requires more study [67] [68] |
| Ideal Application | DNA-free genome editing, somatic hybridization, fundamental studies on single cells [64] [66] | Rapid transient gene expression studies, high-throughput protoplast transformation [67] | Broad-host-range transformation, organellar genome editing, delivery of diverse biomolecules [67] [68] |
Essential Research Reagents and Experimental Protocols
Successful implementation of direct DNA transfer methods requires careful selection of biological materials and adherence to optimized laboratory protocols. The following table outlines key reagent solutions and their critical functions in the transformation workflow.
Table 2: Key Research Reagent Solutions and Their Functions
| Reagent / Material | Function in Experimental Workflow | Application Across Methods |
|---|---|---|
| Cellulases & Pectinases | Enzyme mixture for digesting cell wall components to isolate protoplasts [64] [69] | Essential for protoplast transformation; used in some tissue preps for electroporation |
| Polyethylene Glycol (PEG) | Chemical fusogen that induces membrane perturbation and facilitates DNA uptake [67] [66] | Primarily used in protoplast transformation |
| Gold / Tungsten Microparticles | Microprojectiles coated with genetic cargo for biolistic delivery (a physical method often compared to these techniques) [1] [68] | Not used in the three focal methods, but a common alternative (Biolistics) |
| Cationic Polymers/Lipids | Form stable complexes with nucleic acids via charge interaction, enhancing stability and uptake [67] [68] | Used in nanoparticle-mediated delivery and some protoplast systems |
| Alginate Hydrogel | Polymer matrix for immobilizing protoplasts during culture, reducing aggregation and improving viability [69] | Primarily used in protoplast regeneration systems |
| MES-Mannitol Buffer (MMC) | Osmotically balanced solution to maintain protoplast stability during and after isolation [69] | Critical for protoplast isolation and transformation |
Detailed Experimental Protocol: Protoplast Regeneration
The following workflow, optimized for Arabidopsis thaliana, highlights the intricate steps required for plant regeneration from transformed protoplasts, a common bottleneck for this method [69].
Figure 1: Protoplast Regeneration Workflow. This diagram outlines the multi-stage process for regenerating whole plants from isolated protoplasts, a technique critical for stable transformation via this method [69].
Detailed Experimental Protocol: Nanoparticle-Mediated Delivery
The application of nanoparticles for gene delivery involves a distinct set of preparation and application steps, as summarized below.
Figure 2: Nanoparticle-Mediated Delivery Workflow. This workflow shows the key steps for delivering genetic cargo using nanoparticles, a method noted for its minimal tissue damage and broad host range [67] [68].
The comparative analysis of protoplast transformation, electroporation, and nanoparticle-mediated delivery reveals a diverse landscape of direct DNA transfer methods, each with distinct advantages and constraints. Protoplast systems offer a high-efficiency, single-cell platform ideal for DNA-free genome editing but are hampered by challenging and genotype-dependent regeneration protocols. Electroporation is a well-established, versatile tool for transient studies, yet it requires careful optimization and can impact cell viability. Nanoparticle-mediated techniques emerge as a promising frontier, enabling passive delivery with minimal tissue damage and potential application across a wide range of species, though the technology is still maturing.
The choice of method is not one-size-fits-all but should be guided by the specific research objectives. For rapid transient expression assays or studies in easily regenerable models, protoplast-based systems or electroporation may be optimal. For the modification of recalcitrant species or when seeking to avoid tissue culture, nanoparticle delivery holds significant promise. As plant synthetic biology and precision breeding continue to advance, the integration of these direct delivery methods with cutting-edge tools like CRISPR-Cas9 will be instrumental in developing the next generation of improved crops [64] [65]. Future advancements will likely focus on overcoming the primary limitation of each technique: enhancing protoplast regeneration efficiency, standardizing electroporation parameters for diverse tissues, and developing robust, universal protocols for nanoparticle applications.
Breaking the Bottleneck: Strategies to Enhance Efficiency and Overcome Recalcitrance
Plant genetic transformation serves as a foundational tool for gene function research and crop improvement, yet its application remains constrained by low efficiency and strong genotype dependency across many species [25] [17]. The process typically involves two critical steps: callus formation from explants on auxin-rich callus-inducing medium (CIM), followed by shoot regeneration after transfer to cytokinin-enriched shoot-inducing medium (SIM) [25]. The efficiency of these regeneration stages varies dramatically between species and genotypes, creating a major bottleneck for functional genomics and precision breeding [25] [1].
Developmental regulators (DRs)—key transcription factors controlling cell fate—have emerged as powerful molecular tools to overcome these limitations [70] [71]. By reprogramming somatic cells toward embryogenic or meristematic states, DRs can significantly enhance regeneration capacity in recalcitrant species [54] [72]. This review provides a comparative analysis of four major DR classes—WOX, BBM, WUS, and GRF-GIF—evaluating their molecular mechanisms, transformation efficiency gains, and practical applications in plant biotechnology.
Molecular Mechanisms and Regulatory Networks
WOX Transcription Factors
WUSCHEL-related homeobox (WOX) transcription factors regulate stem cell fate in meristems and are crucial for pluripotency acquisition during callus formation [25] [17]. WOX proteins function by modulating auxin biosynthesis and cytokinin responsiveness, both essential for establishing cellular pluripotency [25]. For example, in apple, MdWOX11 binds the promoter of MdCKX5 to induce its expression, leading to cytokinin degradation and subsequent promotion of adventitious shoot formation [25] [17].
BBM and WUS Partnership
BABY BOOM (BBM), an AP2/ERF domain transcription factor, acts as a key activator of cell proliferation and morphogenesis during somatic embryogenesis [25] [17]. WUSCHEL (WUS), a homeodomain protein, serves as a master regulator of embryogenic and meristematic stem cells [73] [70]. These factors regulate a complex network involving hormone signaling and embryonic competence factors. BBM and WUS transcriptionally regulate LEAFY COTYLEDON1 (LEC1), LEC2, and AGAMOUS-LIKE15 (AGL15) to enhance embryonic competence during somatic embryogenesis [70]. Their functions are intricately linked to hormone signaling networks, with WUS positively regulating cytokinin signaling while BBM enhances cell sensitivity to auxin [70] [71].
GRF-GIF Complexes
GROWTH-REGULATING FACTORS (GRFs) are sequence-specific DNA-binding transcription factors that form functional complexes with GRF-INTERACTING FACTORS (GIFs) to regulate plant growth and developmental processes [73] [25]. GRFs regulate the transition between stem cells to transit-amplifying cells and callus proliferation during organ development, while GIFs boost the transcriptional activity of GRFs [25]. The GRF-GIF complex promotes cell proliferation and plant regeneration, with chimeric GRF-GIF fusion proteins showing enhanced activity in stimulating regeneration [25] [54].
The following diagram illustrates the core regulatory networks governed by these developmental regulators:
Figure 1: Core Regulatory Networks of Key Developmental Regulators. WOX factors primarily influence hormone signaling pathways; BBM activates embryonic competence factors; GRF-GIF complexes drive cell cycle progression. All pathways converge to enhance plant regeneration and genetic transformation efficiency.
Comparative Performance Analysis
Transformation Efficiency Across Species
The application of developmental regulators has demonstrated significant improvements in transformation efficiency across diverse plant species, particularly in previously recalcitrant genotypes. The table below summarizes quantitative data from key studies:
Table 1: Comparative Transformation Efficiency Enhanced by Developmental Regulators
| Developmental Regulator | Species | Genotype | Baseline Efficiency | Enhanced Efficiency | Reference |
|---|---|---|---|---|---|
| TaWOX5 | Wheat (Triticum aestivum) | Jimai22 (recalcitrant) | 5.8% | 55.4% | [25] [17] |
| TaWOX5 | Wheat (Triticum aestivum) | CB037 & Fielder | Not specified | 94.5% | [25] [17] |
| ZmBBM-ZmWUS2 | Maize (Zea mays) | Recalcitrant inbred lines | ~0% | Significant improvement | [73] [70] |
| GRF4-GIF1 | Wheat (Triticum aestivum) | Tetraploid wheat | 2.5% | 63.0% | [54] |
| GRF4-GIF1 | Wheat (Triticum aestivum) | Hexaploid wheat | 12.7% | 61.8% | [54] |
| ZmWIND1 | Maize (Zea mays) | Xiang249 | Not specified | 37.5% (from baseline) | [54] |
| rZmGOLDEN2 | Rice (Oryza sativa) | Four varieties | Variable | 18.1-96.7% regeneration | [54] |
Key Advantages and Limitations
Each developmental regulator class offers distinct advantages and presents specific challenges for practical application:
Table 2: Functional Characteristics of Developmental Regulator Classes
| DR Class | Primary Function | Key Advantages | Major Limitations | Optimal Application |
|---|---|---|---|---|
| WOX | Pluripotency acquisition, meristem maintenance | Less genotype dependency, visual screening markers | Underexplored in many species | Recalcitrant cereal varieties |
| BBM-WUS | Somatic embryogenesis, cell proliferation | Enables transformation of mature tissues, broad species applicability | Pleiotropic effects, developmental abnormalities | Combination approaches with inducible systems |
| GRF-GIF | Cell proliferation, regeneration enhancement | Enables marker-free selection, wide regulatory scope | Requires fusion proteins for optimal effect | Dicot transformation, rapid regeneration systems |
Experimental Protocols and Workflows
BBM-WUS Mediated Transformation
The combined application of BBM and WUS has proven particularly effective for transforming recalcitrant monocot species. The following workflow illustrates a standardized protocol using maize as a model system:
Figure 2: BBM-WUS Mediated Transformation Workflow. Key strategies to mitigate pleiotropic effects include tissue-specific promoters, Cre-lox excision systems, and "altruistic" transformation approaches with transient WUS expression.
GRF-GIF Chimera Implementation
GRF-GIF fusion proteins have demonstrated remarkable efficacy in enhancing regeneration frequency. The experimental protocol typically involves:
Vector Construction: Create GRF-GIF chimeric genes using flexible peptide linkers, driven by constitutive or regeneration-specific promoters [25] [54].
Plant Transformation: Introduce constructs via Agrobacterium-mediated transformation or particle bombardment [54].
Regeneration Optimization: Culture explants on auxin-only medium, as GRF-GIF expression often eliminates cytokinin requirements for shoot regeneration [54].
Selection: Identify transformants using visual markers (e.g., GFP) or antibiotic/herbicide resistance, with the advantage of potentially selecting transgenic plants without antibiotic markers in some systems [54].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for Developmental Regulator Studies
| Reagent / Material | Function | Example Applications |
|---|---|---|
| ZmPLTP promoter | Drives BBM expression in tissue-specific manner | Maize transformation [25] [70] |
| ZmAxig1 promoter | Auxin-inducible WUS expression | Temporal control of morphogenic genes [25] [70] |
| GRF4-GIF1 fusion construct | Enhances regeneration capacity | Wheat, soybean transformation [25] [54] |
| CRE recombinase system | Excises morphogenic genes after transformation | Prevents pleiotropic effects in transgenic plants [25] [70] |
| Ternary vector system | Enhances T-DNA transfer efficiency | Improves transformation in recalcitrant species [70] |
The strategic application of developmental regulators represents a paradigm shift in plant genetic transformation, particularly for recalcitrant species. WOX, BBM-WUS, and GRF-GIF each offer distinct mechanisms for enhancing regeneration efficiency, with documented success across numerous plant families.
Future developments will likely focus on several key areas: (1) refining tissue-specific and inducible expression systems to minimize pleiotropic effects; (2) exploring synergistic combinations of DR classes for additive benefits; (3) adapting these systems to emerging technologies like nanoparticle-mediated delivery and viral vectors for tissue culture-free transformation [54]. As the molecular mechanisms underlying plant regeneration continue to be elucidated, the precision and efficacy of DR-based transformation strategies will undoubtedly expand, further breaking down the barriers to genetic improvement in recalcitrant species.
For researchers embarking on DR-mediated transformation, the selection of an appropriate regulator should be guided by target species, available explant types, and desired transformation workflow. The experimental data and protocols presented herein provide a foundation for developing optimized transformation systems tailored to specific research needs.
Engineering Hormonal Pathways to Boost Callus Formation and Shoot Regeneration
Plant genetic transformation is a cornerstone of modern crop improvement, yet its efficiency is often bottlenecked by the regeneration of whole plants from transformed cells. Central to this process is the precise manipulation of hormonal pathways to induce callus formation and subsequent shoot regeneration. The regenerative capacity of plant cells, known as totipotency, allows a single cell to regenerate an entire plant, but this potential varies significantly across species and genotypes, with many commercially important crops being notoriously recalcitrant [74] [75].
This guide provides a comparative analysis of two dominant strategies for enhancing regeneration: the application of exogenous plant growth regulators and the engineering of endogenous hormonal signaling pathways. We objectively evaluate their performance based on regeneration efficiency, species applicability, and practical implementation within genetic transformation workflows. Supporting experimental data is synthesized to inform researchers and drug development professionals in selecting and optimizing methods for their specific plant systems.
Hormonal Control of Regeneration: Core Principles and Pathways
The Foundational Biphasic Process
Plant regeneration in vitro typically follows a biphasic process. Initially, explants are cultured on an auxin-enriched callus-inducing medium (CIM) to form a pluripotent callus—a mass of undifferentiated cells. Subsequently, this callus is transferred to a shoot-inducing medium (SIM), often with a higher cytokinin-to-auxin ratio, to initiate de novo shoot organogenesis [76].
The Critical Hormonal Balance
The fate of plant cells in vitro is predominantly governed by the interplay of two key hormones:
- Auxins, such as IAA, IBA, and NAA, are crucial for inducing cell dedifferentiation and root formation. They are often termed "rooting hormones" [75].
- Cytokinins, including BAP, kinetin, and zeatin, promote cell division and shoot initiation, earning them the name "shooting hormones" [75].
The ratio of these hormones is a critical determinant:
- A high auxin-to-cytokinin ratio favors root formation.
- A high cytokinin-to-auxin ratio encourages shoot formation.
- Intermediate levels of both often lead to callus proliferation [75].
Table 1: Key Hormones and Their Primary Roles in Plant Regeneration
| Hormone | Class | Primary Function in Regeneration | Commonly Used Types |
|---|---|---|---|
| Auxin | Rooting Hormone | Induces callus formation, root initiation, and cell elongation [75]. | 2,4-D, NAA, IBA, IAA [75] [77] |
| Cytokinin | Shooting Hormone | Promotes cell division and shoot regeneration [75]. | BAP, Kinetin, Zeatin, TDZ [75] [78] |
| Brassinosteroids | Growth Promoter | Enhances callus growth and somatic embryogenesis; regulates cellular metabolism [74]. | Brassinolide (BL) [74] |
| Jasmonic Acid | Stress Hormone | Modulates wound response and can influence regeneration fate [79]. | Jasmonic Acid [79] |
Comparative Analysis of Hormonal Engineering Strategies
Strategy 1: Optimization of Exogenous Hormone Applications
The conventional and most widely used approach involves optimizing the types, concentrations, and combinations of hormones added to the culture medium.
Experimental Protocol for Hormone Optimization
A typical protocol for optimizing exogenous hormones, as demonstrated in Gladiolus, involves:
- Explant Preparation and Sterilization: Use sterile explants such as elongated mother corm sprouts. Surface sterilize with ethanol and sodium hypochlorite [77].
- Callus Induction Medium (CIM): Culture explants on MS medium supplemented with auxins like 2,4-D (2 mg/L) and NAA (2 mg/L), often combined with a cytokinin like BAP (1 mg/L) to induce callus formation [77].
- Callus Maintenance: Subculture callus onto a medium with a lower auxin concentration (e.g., 0.5 mg/L 2,4-D) supplemented with antioxidants (e.g., 150 mg/L ascorbic acid) to control phenolic browning during long-term culture [77].
- Shoot Regeneration Medium (SIM): Transfer callus to MS medium containing a high cytokinin-to-auxin ratio, such as BAP (2 mg/L) and Kinetin (2 mg/L) with a low level of NAA (0.25 mg/L), to promote shoot regeneration [77].
- Rooting and Acclimatization: Regenerated shoots are transferred to a rooting medium, often containing auxins like IAA, and subsequently acclimatized in a greenhouse [77].
Performance and Comparative Data
This strategy's effectiveness is highly species-specific. The table below compares optimized hormone combinations for different species.
Table 2: Comparative Performance of Exogenous Hormone Applications Across Species
| Plant Species | Optimal Hormone Combination for Shoot Regeneration | Reported Regeneration Efficiency | Key Transcription Factors Activated |
|---|---|---|---|
| Arabidopsis thaliana | CIM: Auxin (e.g., 2,4-D); SIM: Cytokinin (e.g., BAP) [76] | Well-established high efficiency | WUS, PLT, LBD family [76] [80] |
| Chinese fir (Cunninghamia lanceolata) | 50 mg/L 6-BA + 0 mg/L NAA + 30 mg/L IBA for callus; 50 mg/L 6-BA + 10 mg/L NAA for bud germination [81] | Callus initiation rate up to 90% [81] | Not specified in study |
| Garnem (G × N15) rootstock | 1.02 mg/L BAP + 0.098 mg/L IBA (optimized via ANN-GA modeling) [78] | Proliferation rate of 10.53 shoots per explant [78] | Not specified in study |
| Gladiolus cultivars | 2 mg/L BAP + 2 mg/L Kin + 0.25 mg/L NAA [77] | 95.55% regeneration; 39.44 shoots per explant [77] | Genetic stability confirmed via ISSR markers [77] |
| Brettschneidera sinensis | 1.0 mg/L 6-BA + 0.1 mg/L NAA for shoot regeneration [74] | High shoot regeneration rate reported [74] | Genetically stable per ISSR/RAPD [74] |
Advantages: This method is technically accessible, highly tunable, and does not require genetic modification of the plant material. Limitations: Efficiency is highly genotype-dependent, risk of somaclonal variation with long-term culture, and optimization can be time-consuming [77].
Strategy 2: Engineering Endogenous Signaling Pathways
A more advanced strategy involves genetically engineering components of key signaling pathways to boost the plant's innate regenerative capacity.
Key Signaling Pathways and Engineering Targets
- The CLE-CLV1/BAM1 Module: The CLAVATA3/EMBRYO SURROUNDING REGION-RELATED (CLE) peptides, particularly CLE1-CLE7 and CLE9/10, act as negative regulators of shoot regeneration. They are perceived by receptors CLV1 and BAM1, which restrict the expression of WUSCHEL (WUS)—a master regulator of shoot stem cell identity. Mutants in cle1-7 or clv1/bam1 show enhanced shoot regeneration capacity [76].
- The REF1-PORK1-WIND1 Positive Feedback Loop: The REGENERATION FACTOR1 (REF1) peptide, upon binding its receptor PORK1, activates the transcription factor WOUND-INDUCED DEDIFFERENTIATION 1 (WIND1). WIND1, in turn, promotes callus formation and shoot regeneration and also binds to the PRP (REF1 precursor) promoter to amplify its own signal, creating a positive feedback loop that enhances regenerative capacity [76].
- The RALF33-FER Module in Root Regeneration: The RAPID ALKALINIZATION FACTOR 33 (RALF33) peptide accumulates at wound sites and inhibits the receptor kinase FERONIA (FER), leading to the activation of the TPR4-ERF115 module, which promotes root regeneration [76].
Experimental Protocol for Synthetic Pathway Engineering
A groundbreaking tissue-culture-free protocol (Texas Tech University, 2025) leverages this knowledge [82]:
- Genetic Construction: Create a synthetic gene cassette combining a wound-inducible promoter with key regulatory genes, such as WIND1 (to trigger cell reprogramming) and the isopentenyl transferase (IPT) gene (to locally produce cytokinin).
- Plant Transformation: Introduce this cassette into the plant genome using Agrobacterium-mediated transformation or other methods.
- Activation and Regeneration: Wounding the transformed plant (e.g., leaf puncture) activates the synthetic circuit. The local expression of WIND1 and IPT initiates a regeneration cascade, leading to the direct formation of gene-edited shoots from the wound site, bypassing the callus stage [82].
Performance and Comparative Data
This emerging strategy shows promise for overcoming recalcitrance.
Table 3: Performance of Engineered Endogenous Pathways
| Engineering Target / Approach | Effect on Regeneration | Reported Efficiency / Application |
|---|---|---|
| CRISPR knockout of CLE1-7 | Negative regulator removal; enhances shoot regeneration [76] | Increased number of adventitious shoots in Arabidopsis [76] |
| Overexpression of REF1 peptide | Positive regulator addition; enhances callus formation and shoot regeneration [76] | Improved regeneration and transformation in soybean, wheat, and maize [76] |
| Synthetic WIND1-IPT circuit | Bypasses tissue culture; induces shoots directly from wounds [82] | Success in tobacco, tomato, and soybean; higher success rates in tobacco/tomato [82] |
| Mutation in ralf33 | Reduces root regeneration capacity [76] | Lower regeneration rates after root tip resection [76] |
Advantages: Can overcome species-specific recalcitrance, enables tissue-culture-free transformation, and can lead to higher regeneration efficiencies in optimized systems. Limitations: Requires genetic transformation capability, potential for pleiotropic effects, and regulatory considerations for genetically modified organisms (GMOs).
The Impact of Environmental and Other Factors
The hormonal regulation of regeneration does not occur in a vacuum. Environmental and other internal factors play a critical modulatory role.
- Water Availability: Recent research reveals that local water availability at the wound site is a decisive factor in regeneration fate. High water availability promotes the formation of auxin response maxima that drive root regeneration, while low water availability favors callus formation through cambium-related pathways. This process is mediated by water's influence on stress hormones like ethylene and jasmonic acid, which in turn modify auxin transport [79].
- Light Quality: Light acts as a key environmental regulator. Blue and red light spectra are known to promote adventitious shoot regeneration, while light intensity and photoperiod exert their effects by influencing hormonal activity and photosynthesis [74].
- Metabolites and Oxidative Stress: Metabolites in the phenylpropanoid and flavonoid biosynthesis pathways can accumulate in explants from older trees, leading to lignification of the callus and inhibition of root formation [74]. Oxidative stress from free radicals is also a major contributor to somaclonal variation and reduced regeneration efficiency [77].
The Scientist's Toolkit: Essential Research Reagents
Table 4: Key Reagents for Hormonal Pathway Research in Plant Regeneration
| Research Reagent / Solution | Function and Application in Regeneration Studies |
|---|---|
| Synthetic CLE Peptides | Used to experimentally suppress shoot regeneration and study the role of this pathway; applied exogenously in a dose-dependent manner [76]. |
| Synthetic REF1 Peptide | Applied exogenously to enhance callus formation and shoot regeneration in wild-type plants and to rescue regenerative defects in mutants [76]. |
| Activated Charcoal & Antioxidants | Used in culture media (e.g., 150 mg/L ascorbic acid, 500 mg/L activated charcoal) to adsorb inhibitory phenolic compounds exuded by explants, improving callus health and regeneration [77]. |
| Murashige and Skoog (MS) Medium | The foundational basal medium for most plant tissue culture protocols, providing essential macro and micronutrients [81] [77]. |
| WOUND-Inducible Promoters | Genetic tools used to drive the expression of regeneration-enhancing genes (e.g., WIND1, IPT) specifically at wound sites, mimicking natural regeneration triggers [82]. |
Signaling Pathway Diagrams
Negative Regulation of Shoot Regeneration via CLE Signaling
Positive Enhancement of Regeneration via the REF1 Pathway
Environmental Influence on Regeneration Fate
The comparative analysis presented in this guide reveals a spectrum of strategies for engineering hormonal pathways to boost plant regeneration. The choice of strategy depends heavily on the research goals and target species. Optimizing exogenous hormone applications remains a highly accessible and powerful method for many applications, particularly where GMOs are not desirable. In contrast, engineering endogenous pathways represents the cutting edge, offering a promising solution to the persistent challenge of recalcitrant species and holding the potential to dramatically accelerate the development of transgenic and gene-edited crops.
Future research will likely focus on refining these engineered pathways, identifying new regulatory targets, and combining these approaches with an improved understanding of environmental influences to develop universal and highly efficient plant transformation systems.
Plant genetic transformation is a cornerstone of modern crop improvement and functional genomics. For over three decades, the gene gun (biolistic delivery) has been a vital tool, particularly for species and tissue types recalcitrant to Agrobacterium-mediated transformation. However, its widespread application has been hampered by long-standing challenges of inefficiency, inconsistency, and tissue damage [21] [46].
A recent breakthrough from an interdisciplinary team has addressed these limitations at a fundamental level. Through advanced computational modeling and plant biotechnology, researchers have developed the Flow Guiding Barrel (FGB), a simple yet transformative device that enhances the performance of the standard gene gun by up to 22-fold [21]. This guide provides a comparative analysis of the FGB's performance against conventional biolistic and other transformation methods, detailing the experimental data and protocols that validate its efficacy.
The Flow Guiding Barrel (FGB): A Paradigm Shift in Biolistics
Fundamental Innovation and Mechanism of Action
The core innovation of the FGB lies in its re-engineering of the gas and particle flow dynamics within the gene gun. Computational Fluid Dynamic (CFD) simulations of the widely used Bio-Rad PDS-1000/He system revealed a critical design flaw: the small aperture of the internal barrel creates a restrictive bottleneck [21] [46].
This results in:
- Significant particle loss (only ~21% of loaded particles reach the target)
- Disrupted, diffusive helium flow
- Reduced particle velocity and pressure
- Uneven distribution on target tissue [21]
The FGB, a 3D-printed replacement for the gun's internal spacer rings, is engineered to systematically optimize this flow. It enables a uniform laminar flow pattern, directing nearly 100% of the loaded microprojectiles toward the target with higher velocity and over a four-times larger area [21]. The table below summarizes this fundamental improvement.
Table 1: Fundamental Performance Comparison: Conventional Barrel vs. Flow Guiding Barrel (FGB)
| Performance Parameter | Conventional Barrel | Flow Guiding Barrel (FGB) | Improvement Factor |
|---|---|---|---|
| Particle Delivery Efficiency | ~21% of loaded particles | ~100% of loaded particles | ~4.8-fold |
| Flow Pattern | Inconsistent, diffusive | Uniform, laminar | N/A |
| Target Coverage Area | 1.77 cm² | 7.07 cm² | 4-fold |
| Particle Penetration Depth | Single peak at 7.5 µm | Deeper penetration, peaks at 5-7.5 µm & 22.5-25 µm | 2.3-fold max depth |
| Simulated Particle Velocity | Baseline | Twice the velocity | 2-fold |
Experimental Validation and Performance Data
The FGB has been rigorously tested across a wide range of applications and plant species. The following table synthesizes the key quantitative outcomes from these experiments, demonstrating its superior performance.
Table 2: Experimental Results of FGB Performance Across Various Applications
| Application / Target Tissue | Delivered Cargo | Key Performance Metric | Conventional System | FGB System | Improvement Factor |
|---|---|---|---|---|---|
| Onion Epidermis | GFP-DNA (22 ng) | Transient transfection (fluorescent cells) | 153 cells | 3,351 cells | 22-fold [21] |
| Onion Epidermis | CRISPR-Cas9 RNP | Gene editing efficiency (F3'H gene) | Baseline | 6.6% editing (NGS) | 4.5-fold [21] |
| Maize Seedlings | SCMV-CS1-GFP Virus | Viral infection rate | 5% | 83.5% | 17-fold [21] |
| Soybean Seedlings | SMV-GFP Virus | Viral infection rate | 66% | 100% | 1.5-fold [21] |
| Maize B104 Immature Embryos | pCBL101-mCherry | Stable transformation frequency | Baseline | >10-fold increase [21] | >10-fold |
| Wheat Shoot Apical Meristems | CRISPR-Cas12a | Heritable editing efficiency (T0/T1) | Baseline (3 bombardments) | 2-fold increase (1 bombardment) | 2-fold (with fewer shots) [21] |
Comparative Analysis of Plant Transformation Methods
While biolistics is a powerful direct delivery method, other established techniques include Agrobacterium-mediated transformation and protoplast-mediated transformation [83]. The advent of the FGB significantly shifts the comparative landscape, enhancing the position of biolistic delivery.
Table 3: Comparative Overview of Major Plant Transformation Methods
| Transformation Method | Key Mechanism | Key Advantages | Key Limitations | Impact of FGB |
|---|---|---|---|---|
| Biolistic (Conventional) | High-velocity microprojectiles | • Species/tissue independent• Delivers DNA, RNA, RNP [21] | • Low efficiency & consistency• High tissue damage• Complex transgene inserts [21] [46] | Directly addresses core limitations |
| Agrobacterium-mediated | Natural DNA transfer from bacteria | • High efficiency• Typically low-copy, clean inserts [83] | • Narrow host range for many strains• Cannot deliver RNPs [21] | N/A |
| Protoplast-mediated | Direct DNA uptake into naked cells | • High-throughput for transient expression [83] | • Requires cell wall regeneration• Not genotype-independent [83] | N/A |
| Biolistic (with FGB) | Optimized particle flow dynamics | • All conventional advantages PLUS:• >10x higher efficiency• Reduced tissue damage• Higher consistency [21] [46] | • Still requires specialized equipment | Mitigates key disadvantages of conventional biolistics |
Detailed Experimental Protocols Featuring the FGB
To facilitate adoption and replication, here are the detailed methodologies for key experiments validating the FGB.
Protocol 1: Transient Transformation and Genome Editing in Onion Epidermis
This protocol is used for rapid assessment of DNA, protein, or ribonucleoprotein (RNP) delivery [21].
- Microcarrier Preparation: Suspend 1µm gold particles in 100% ethanol. Coat particles with plasmid DNA (e.g., pLMNC95 for GFP) or pre-assembled CRISPR-Cas9 RNP complexes. For protein delivery, coat with FITC-labeled BSA (FITC-BSA).
- Gene Gun Setup: Install the FGB device into the bombardment chamber of a Bio-Rad PDS-1000/He system, replacing the standard spacer rings.
- Target Preparation: Peel the inner epidermis of an onion bulb and place it adaxial side up on solid medium in a Petri dish.
- Bombardment Parameters: Set the helium pressure to 450 psi and the target distance to 6 cm. Conduct a single bombardment per sample.
- Incubation and Analysis: Incubate samples in the dark at 25°C for 24-48 hours.
- For GFP/FITC-BSA: Analyze using fluorescence microscopy and count fluorescent spots.
- For CRISPR RNP: Extract genomic DNA from bombarded tissue and analyze target gene editing efficiency via next-generation sequencing (NGS).
Protocol 2: Stable Transformation of Maize Immature Embryos
This protocol demonstrates the FGB's impact on stable transformation, a critical application for crop engineering [21].
- Explant Preparation: Isolate immature embryos (1.5-2.0 mm) from maize inbred line B104 10-12 days after pollination.
- Pre-culture: Place approximately 100 embryos scutellum-side up on osmotic medium. This high throughput is enabled by the FGB's larger bombardment area, a significant increase from the 30-40 embryos used with conventional barrels.
- Microcarrier Preparation: Coat gold particles with the plasmid vector of interest (e.g., pCBL101-mCherry for visual selection).
- Bombardment: Bombard embryos using the FGB at a helium pressure of 650 psi and a target distance of 9 cm.
- Selection and Regeneration: Transfer embryos to callus induction medium with a selective agent (e.g., hygromycin). Regenerate plantlets from resistant calli and transfer to soil for molecular confirmation and seed production.
The Scientist's Toolkit: Essential Research Reagents & Materials
Table 4: Key Reagents and Materials for FGB-Based Transformation
| Item | Function/Description | Example Application |
|---|---|---|
| Bio-Rad PDS-1000/He System | Standard gene gun device for biolistic delivery. | Platform for FGB installation and use [21]. |
| Flow Guiding Barrel (FGB) | 3D-printed device optimizing gas/particle flow. | Core innovation enhancing all bombardment efficiency [21] [46]. |
| Gold Microcarriers (0.6-1.0 µm) | Inert particles that carry biological cargo into cells. | Coated with DNA, RNA, or proteins for delivery [21]. |
| CRISPR-Cas9/12a RNP Complexes | Pre-assembled complexes of Cas protein and guide RNA. | Enables DNA-free, precise genome editing with reduced off-target effects [21]. |
| Plasmid Vectors with Reporter Genes | e.g., pLMNC95 (GFP), pCBL101 (mCherry). | Visual markers for rapid assessment of transient and stable transformation efficiency [21]. |
| Spermidine (Precipitating Agent) | Helps adsorb DNA onto the surface of microcarriers. | Standard component in microcarrier coating procedure [21]. |
Mechanism of FGB Action: A Visual Workflow
The following diagram illustrates the logical workflow of the problem identification, solution development, and performance outcome that characterizes the FGB innovation.
The Flow Guiding Barrel represents a landmark optimization in biolistic technology. By addressing fundamental flow dynamics issues, it transforms a decades-old tool, achieving order-of-magnitude improvements in efficiency across a wide spectrum of applications—from transient protein expression to stable crop transformation and DNA-free genome editing [21]. For researchers and drug development professionals, the FGB enables higher throughput, greater reliability, and expanded capabilities in plant genetic engineering. It is particularly impactful for engineering recalcitrant species and for advancing transgene-free CRISPR editing, directly addressing regulatory and public concerns [21] [46]. This innovation firmly repositions biolistic delivery as a highly efficient and versatile platform within the modern plant synthetic biology toolkit.
Plant genetic transformation is a cornerstone of modern crop improvement, enabling functional genomics research and the development of novel traits. However, the efficiency of these technologies has long been constrained by a significant bottleneck: strong genotype dependence. Conventional transformation methods often succeed only in a limited number of laboratory-friendly model genotypes, leaving many elite, commercially valuable cultivars recalcitrant to genetic modification [84] [25]. This limitation severely restricts the application of advanced breeding technologies, such as genetic engineering and gene editing, for the improvement of a wide range of crop species.
The core of the problem lies in the intricate process of plant regeneration from transformed cells. Traditional methods rely on tissue culture, a process where the efficiency of callus formation and subsequent shoot regeneration is highly influenced by a plant's genetic makeup [25]. Consequently, researchers have pursued innovative strategies to overcome this biological barrier. This guide provides a comparative analysis of the leading genotype-independent transformation strategies, evaluating their operational principles, experimental protocols, and relative performance across diverse plant species.
Comparative Analysis of Genotype-Independent Transformation Strategies
The following table summarizes the key features, advantages, and limitations of the primary strategies employed to achieve genotype-independent transformation.
Table 1: Overview of Major Genotype-Independent Transformation Strategies
| Strategy | Core Principle | Key Advantages | Major Limitations | Representative Efficiency |
|---|---|---|---|---|
| Developmental Regulator-Assisted | Overexpression of morphogenic genes (e.g., BBM, WUS) to enhance regeneration [84] [85]. | Dramatically improved efficiency in recalcitrant genotypes; shorter transformation time [84] [25]. | Potential for developmental abnormalities and sterility in T0 plants [84] [25]. | Up to 94.5% in wheat using TaWOX5 [25]. |
| In Planta Transformation | Direct transformation of intact plants or explants with minimal to no tissue culture [19] [86]. | Bypasses tissue culture; technically simple, affordable, and less genotype-dependent [19] [86]. | Often lower transformation frequency; can be species-specific [86]. | 14.3% - 45.0% in chickpea and pigeon pea [86]. |
| Altruistic Transformation | Transient expression of morphogenic genes in neighboring cells to stimulate somatic embryogenesis for transformed cells [25]. | High efficiency without integrating morphogenic genes into the final plant [25]. | Requires careful optimization of Agrobacterium strain ratios [25]. | 19.5% (avg.) in maize, a 2.5-fold increase over conventional methods [25]. |
| Novel Explant Utilization | Use of alternative, highly regenerative tissues like internodal segments [87]. | Amenable to high-throughput transformation; applicable to elite germplasm [87]. | Explant availability and quality can be variable. | Successful transformation of 12 elite canola genotypes, previously recalcitrant [87]. |
Experimental Protocols and Workflows
Developmental Regulator-Assisted Transformation
This strategy involves the co-transformation of the gene of interest with key developmental regulatory genes that promote cell proliferation and regeneration.
Table 2: Key Morphogenic Genes and Their Functions in Plant Regeneration
| Gene | Gene Family | Primary Function in Regeneration | Commonly Used Promoters |
|---|---|---|---|
| BbM | AP2/ERF | Master regulator of cell proliferation and somatic embryogenesis [25]. | Maize Ubiquitin (ZmUbi), ZmPLTP [84] [25]. |
| WUS2 | WOX | Master regulator of stem cell fate and embryogenic stem cells [25]. | Nopaline synthase (NOS), ZmAxig1 [84] [25]. |
| WOX5 | WOX | Key regulator of pluripotency acquisition in callus [25]. | Constitutive promoters (e.g., 35S, Ubiquitin) [25]. |
Detailed Workflow: The diagram below outlines the key protocol for developmental regulator-assisted transformation using morphogenic genes.
The "Altruistic" Transformation System
This innovative system relies on the transient, non-inherited expression of morphogenic genes to stimulate regeneration in a cell-non-autonomous manner.
Detailed Workflow: The altruistic transformation process uses a mixed Agrobacterium approach to stimulate somatic embryogenesis in neighboring cells.
In PlantaTransformation Protocol
This genotype-independent method bypasses tissue culture by directly transforming germinating seeds.
Detailed Workflow: The in planta transformation method uses germinating seeds for direct Agrobacterium-mediated transformation without tissue culture.
Quantitative Efficiency Data Across Species
The following table compiles experimental transformation efficiency data for different strategies across various crop species, demonstrating their genotype-independent potential.
Table 3: Comparative Transformation Efficiencies Across Species and Methods
| Crop Species | Genotype | Method | Key Genes/Explant | Transformation Efficiency | Reference |
|---|---|---|---|---|---|
| Wheat | Fielder (model) | Developmental Regulator | TaWOX5 | Up to 94.5% | [25] |
| Wheat | Jimai22 (recalcitrant) | Developmental Regulator | TaWOX5 | 5.8% → 55.4% | [25] |
| Maize | Recalcitrant lines | Altruistic System | ZmWUS2 + lethal gene | Avg. 19.5% (2.5x increase) | [25] |
| Chickpea | HC-1 | In Planta | Germinating seeds | 40.9% | [86] |
| Pigeon Pea | Manak | In Planta | Germinating seeds | 45.0% | [86] |
| Canola | 12 elite lines | Novel Explant | Internodal segments | Successful in all 12 genotypes | [87] |
| Sorghum | Recalcitrant | BBM/WUS | ZmBBM + ZmWUS2 | Enhanced frequency | [84] [25] |
The Scientist's Toolkit: Essential Research Reagents
Table 4: Key Reagent Solutions for Genotype-Independent Transformation
| Reagent / Material | Function / Application | Examples & Notes |
|---|---|---|
| Morphogenic Gene Constructs | Enhance regeneration competence in transformed cells. | BBM, WUS2, WOX5; use with inducible systems or tissue-specific promoters to avoid pleiotropic effects [84] [25]. |
| Agrobacterium Strains | Biological vector for T-DNA delivery. | LBA4404, EHA105; strain choice can impact efficiency in different species [86] [87]. |
| Novel Explant Types | Bypass recalcitrance by using highly regenerative tissues. | Internodal segments [87], shoot apical meristems (SAM) [19], germinating seeds [86]. |
| Selection Agents | Selective growth of transformed cells. | Antibiotics (spectinomycin), herbicides; in planta methods may use post-selection screening [86] [87]. |
| Nanoparticles | Physical delivery of biomolecules, protecting cargo and bypassing cell wall. | Magnetic nanoparticles (MNPs), carbon nanotubes; emerging tool for species recalcitrant to Agrobacterium [85] [23]. |
The pursuit of genotype-independent transformation has yielded a diverse toolkit of strategies, each with distinct mechanisms and applications. Developmental regulator-assisted methods offer the highest efficiencies by directly reprogramming cell fate but require careful handling to avoid adverse phenotypes. The altruistic transformation system provides an elegant solution to this problem by spatially separating the regeneration stimulus from the stable transformation event. Meanwhile, in planta methods present the most accessible and technically simple path for many laboratories, bypassing tissue culture entirely.
The choice of strategy is inherently species- and context-dependent. For high-throughput transformation in major cereals, developmental regulators and altruistic systems currently lead the field. For minor crops, legumes, or resource-limited settings, robust in planta protocols offer a viable and rapid alternative. As these technologies continue to mature and converge with genome editing tools, they promise to unlock the full potential of precision breeding across the vast diversity of crop species and elite cultivars, finally overcoming the longstanding barrier of genotype dependency.
Plant genetic transformation is a cornerstone of modern crop improvement and functional genomics. However, a significant bottleneck persists: the reliance on complex, time-consuming, and genotype-dependent tissue culture processes [1]. These traditional methods are often inefficient, can induce somaclonal variation, and are unsuitable for many recalcitrant species, particularly perennial and woody crops [88] [89]. In response, the field is advancing on two complementary fronts: the development of tissue culture-free ("in planta") transformation methods and the adoption of visual selection markers to replace antibiotic-based selection. This guide provides a comparative analysis of these innovative approaches, detailing their experimental protocols, efficiencies, and practical applications to help researchers streamline their genetic transformation workflows.
Visual Selection Markers: A Paradigm Shift from Antibiotics
Traditional selection methods rely on selectable marker genes (SMGs) that confer resistance to antibiotics or herbicides. While effective, they face growing regulatory and consumer scrutiny, and their use requires the continuous application of selective agents throughout the tissue culture stage [89] [90]. Visual markers offer a compelling alternative by enabling the direct, non-invasive identification of transformed tissues without the need for toxic compounds.
Table 1: Comparison of Major Visual Selection Marker Systems
| Marker System | Key Components / Gene | Visible Phenotype | Substrate Required? | Primary Advantages | Reported Transformation Efficiencies |
|---|---|---|---|---|---|
| Anthocyanin (MYB10) | Apple MYB10 transcription factor [91] | Red/Purple pigmentation in callus, shoots, and roots | No | Non-toxic; potential health benefits; can replace kanamycin selection [91] | Apple: Red shoots always PCR-positive [91]; Potato: Up to 4x higher anthocyanin content in PCR-positive shoots [91] |
| Betalain (RUBY) | Synthetic construct (CYP76AD1, DODA, Glucosyltransferase) linked by 2A peptides [92] | Vivid red betalain pigment in tissues and seeds | No (uses endogenous tyrosine) | Visible to naked eye; no equipment or substrates needed; effective for monocots and dicots [92] | Effective selection in Arabidopsis and rice tissue culture; allows easy tracking of single-insertion events in seeds [92] |
| GFP/mCherry | Green/Red Fluorescent Protein | Green/Red fluorescence under specific light | Yes (specific light wavelength) | Allows precise cellular localization | N/A in provided results |
| GUS (β-glucuronidase) | uidA gene | Blue precipitate upon staining | Yes (X-Gluc) | Robust and well-characterized histochemical stain | N/A in provided results |
Experimental Protocols for Key Visual Markers
Anthocyanin-Based Selection with MYB10 The mutant allele of the apple MYB10 transcription factor can be used as a visual selectable marker. The protocol involves:
- Vector Construction: The MYB10 gene, including its own promoter and terminator, is cloned into a transformation vector [91].
- Transformation and Regeneration: Standard Agrobacterium-mediated transformation is performed on explants (e.g., apple leaf discs, strawberry runners, potato stem segments). After co-cultivation, explants are transferred to regeneration medium without kanamycin [91].
- Selection and Identification: Putatively transformed tissues are identified visually by their red/purple coloration during callus formation and shoot regeneration. Red shoots can be excised and propagated. Studies show that red shoots are always positive for the transgene, though not all transgenic shoots are intensely red, suggesting a sensitivity trade-off [91].
- Light Conditions: Note that anthocyanin accumulation is light-dependent. For apple, initial callus induction was performed in the dark, but anthocyanin production was more frequent and specific upon transfer to standard light conditions (50 μE) [91].
Betalain-Based Selection with RUBY The RUBY reporter system is a single open reading frame encoding three enzymes (CYP76AD1, DODA, and a glucosyltransferase) required for betalain biosynthesis, connected by self-cleaving 2A peptides [92].
- Vector Construction: The RUBY cassette is placed under a constitutive promoter (e.g., CaMV 35S) or a tissue-specific promoter (e.g., seed-specific At2S3) in the transformation vector [92].
- Transformation: For Arabidopsis, the floral dip method is used. Transformed seeds (T1) are easily distinguished by their dark red color and sown directly [92].
- In Tissue Culture: For species like rice, the construct is transformed into calli. Transformed calli and regenerated plantlets display a vivid red color, allowing for easy visual selection throughout the process without adding substrates or using specialized equipment [92].
The following diagram illustrates the core principle of visual marker systems, using the betalain biosynthesis pathway in the RUBY system as an example.
Tissue Culture-Free (In Planta) Transformation Methods
In planta transformation refers to a heterogeneous group of techniques that directly introduce DNA into intact plants or plant tissues, bypassing or minimizing the need for in vitro tissue culture [19]. These methods are often simpler, faster, more affordable, and less genotype-dependent than conventional methods.
Table 2: Comparison of Major In Planta Transformation Methods
| Method | Key Explant/Tissue | Basic Protocol Summary | Key Advantages | Reported Efficiencies & Applications |
|---|---|---|---|---|
| Floral Dip [19] | Developing flowers & ovules | Dip inflorescences in Agrobacterium suspension; T1 seeds screened. | Simple, no tissue culture; high-throughput. | Gold standard for Arabidopsis; adapted for some grasses and crops like soybean and wheat [19]. |
| Pollen-Based [35] [19] | Pollen grains | Transform pollen via electroporation, magnetofection, or Agrobacterium; use for pollination. | Targets male germline; avoids chimerism. | Success in genera like Brassica, Dianthus, Petunia [19]. |
| Pollen Tube Pathway [1] [19] | Pollen tube | Inject DNA solution into ovary post-pollination; pollen tube delivers DNA. | Bypasses tissue culture; technically simple. | Applied in cotton, melon, soybean, wheat, maize; 2.54% efficiency in Paphiopedilum [1]. |
| Shoot Apical Meristem (SAM) [35] [19] | Meristematic cells | Infect/Inject Agrobacterium into vegetative or embryonic SAM; recover shoots. | Bypasses callus phase; direct regeneration. | Applied in >30 species including chickpea, pigeon pea, cotton; genotype-independent [19]. |
| Mechanical Damage (Cut-Dip-Budding) [93] | Vegetative buds & meristems | Scarify buds, dip in Agrobacterium, recover shoots in vivo. | Minimal sterile culture; applicable to perennials. | Transformation efficiencies: C. stauntonii (51.7%), A. argyi (9.3%), C. morifolium (16.7%) [93]. |
Experimental Protocols for Key In Planta Methods
Floral Dip Transformation This is the benchmark for in planta transformation in Arabidopsis thaliana [19].
- Plant Growth: Grow plants until the primary inflorescence is ~5-10 cm tall, and secondary inflorescences have started to appear.
- Agrobacterium Preparation: Inoculate a culture of Agrobacterium tumefaciens carrying the binary vector and grow to saturation. Pellet the bacteria and resuspend in a 5% sucrose solution, often with a surfactant like Silwet L-77 (~0.02-0.05%).
- Dipping: Submerge the above-ground portions of the plant (focusing on inflorescences) in the bacterial suspension for a few minutes, with gentle agitation.
- Recovery and Seed Harvest: Lay the dipped plants on their side and cover to maintain high humidity for 16-24 hours. Return plants to normal growth conditions. Harvest seeds (T1) once fully dried.
- Screening: Sow T1 seeds under selective conditions (e.g., containing an antibiotic or by visual screening with a marker like RUBY) to identify primary transformants.
Shoot Apical Meristem (SAM) Transformation This method targets the pluripotent cells of the meristem.
- Explant Preparation: Seeds are sterilized and germinated on a sterile medium. Alternatively, vegetative buds from mature plants can be used [93] [19].
- Meristem Exposure/Injury: For seedlings, the meristem is exposed by removing the primary leaves. Mechanical injury by scraping the base of the bud with a sterile blade is a key step to enhance Agrobacterium infection [93].
- Agrobacterium Co-cultivation: The exposed meristems are inoculated with an Agrobacterium suspension, either by vacuum infiltration, direct injection, or simple dipping, and then co-cultivated for 1-3 days.
- Recovery and Regeneration: Unlike tissue culture methods, the infected seedlings or buds are often transferred directly to soil or a minimal medium without prolonged callus induction phases. The transformed cells within the meristem give rise to chimeric shoots. Non-chimeric, transformed shoots (T0) are recovered through selection and subsequent generations (T1) are screened to identify stable transformants.
The following workflow diagram generalizes the key steps for several in planta methods, highlighting their simplified nature compared to traditional tissue culture.
The Scientist's Toolkit: Essential Reagents and Materials
Successful implementation of these simplified workflows relies on a core set of reagents and materials.
Table 3: Research Reagent Solutions for Simplified Transformation Workflows
| Reagent/Material | Function/Purpose | Examples/Specific Notes |
|---|---|---|
| Visual Marker Vectors | Non-invasive selection of transformants without antibiotics. | pMYB10: For anthocyanin-based selection [91]. RUBY vectors: For betalain-based selection in dicots and monocots [92]. |
| Developmental Regulators (DRs) | Enhance transformation and regeneration efficiency, potentially overcoming genotype limitations. | BBM/WUS2: Promote somatic embryogenesis [18]. GRF-GIF: Enhances shoot regeneration, may allow hormone-free regeneration [18]. |
| Agrobacterium Strains | Delivery of T-DNA into plant cells. | Strain specificity is key. A. rhizogenes MSU440 and A4 showed high efficiency in medicinal plant transformation [93]. |
| Surfactants | Lowers surface tension of inoculation medium, improving Agrobacterium penetration into tissues. | Silwet L-77: Critical for floral dip and vacuum infiltration methods [19]. |
| Sucrose Solution | Basis for Agrobacterium resuspension during inoculation; provides osmoticum and energy. | Typically used at 5% concentration for floral dip and other in planta methods [19]. |
The combined use of tissue culture-free transformation methods and visual selection markers represents a powerful strategy to overcome the historical bottlenecks in plant genetic engineering. As detailed in this guide, methods like floral dip, meristem transformation, and the pollen tube pathway offer streamlined, genotype-flexible alternatives to tissue culture. When paired with visible reporters like MYB10 or RUBY, which eliminate the need for antibiotic-based selection, these workflows become more efficient, cost-effective, and accessible. While challenges remain—such as optimizing these protocols for a wider range of perennial and recalcitrant species—the continued development and adoption of these tools are poised to accelerate both basic plant research and precision breeding efforts.
Data-Driven Decision Making: Validating and Comparing Transformation Efficiency
Plant genetic transformation serves as a foundational technique in modern plant biotechnology, enabling functional genomics studies, crop improvement, and the production of valuable recombinant proteins [1] [94]. The efficiency of these transformation processes is typically quantified through three primary metrics: stable transformation frequency, which measures the successful integration of foreign DNA into the plant genome resulting in heritable modifications; transient transformation efficiency, which assesses short-term gene expression without genomic integration; and editing rates, which quantify the success of targeted genome modifications using technologies like CRISPR/Cas9 [94] [54] [95]. These metrics provide crucial benchmarks for comparing and optimizing transformation protocols across diverse plant species and experimental systems.
The selection of an appropriate transformation strategy presents a significant bottleneck in plant biotechnology, particularly for non-model species and recalcitrant crops [19] [54]. This challenge is compounded by the genotype-dependent nature of many transformation systems and the extensive tissue culture requirements that can lead to somaclonal variations [1] [19]. This guide provides a comprehensive, data-driven comparison of current plant transformation methodologies, focusing on quantitative performance metrics across different experimental systems. By synthesizing empirical data from recent studies, we aim to equip researchers with the analytical framework necessary to select optimal transformation strategies for their specific experimental needs, ultimately accelerating progress in plant genetic engineering and genome editing applications.
Comparative Analysis of Transformation Metrics Across Methods
Stable Transformation Frequency
Stable transformation represents the permanent integration of foreign DNA into the plant genome, enabling hereditary transmission of the transgene to subsequent generations [94]. This process typically involves more complex and lengthy procedures than transient transformation but provides enduring genetic modifications essential for long-term trait development and breeding programs.
Table 1: Comparative Stable Transformation Frequencies Across Methods and Species
| Transformation Method | Plant Species | Explants Used | Transformation Frequency | Key Factors Influencing Efficiency | Citation |
|---|---|---|---|---|---|
| Agrobacterium-mediated (Standard) | Maize (inbred lines) | Immature embryos | 16.56%-37.5% | Co-expression of developmental regulators (ZmWIND1) | [54] |
| Agrobacterium-mediated (Standard) | Wheat (varieties) | Immature embryos | 17.5%-96.2% | Expression of TaWOX5; genotype dependence | [54] |
| Agrobacterium-mediated (Standard) | Tetraploid wheat | Immature embryos | 2.5% (control) to 63.0% (optimized) | Co-expression of GRF4-GIF1 fusion protein | [54] |
| Agrobacterium-mediated (Standard) | Hexaploid wheat | Immature embryos | 12.7% (control) to 61.8% (optimized) | Co-expression of GRF4-GIF1 fusion protein | [54] |
| Agrobacterium-mediated (Standard) | Tomato (wild) | Leaf explants | 6- to 12-fold increase | Application of REF1 regeneration factor | [54] |
| Tissue culture-free (Cut-dip-budding) | Cynanchum stauntonii (Medicinal) | Seedlings | 51.7% | Mechanical damage; strain A4 | [93] |
| Tissue culture-free (Cut-dip-budding) | Artemisia argyi (Medicinal) | Seedlings | 9.3% | Mechanical damage; strain MSU440 | [93] |
| Tissue culture-free (Cut-dip-budding) | Chrysanthemum morifolium (Medicinal) | Seedlings | 16.7% | Mechanical damage; strain MSU440 | [93] |
| Floral dip (in planta) | Arabidopsis thaliana | Inflorescence | Highly variable | Developmental stage, surfactant concentration | [19] [94] |
| Pollen-tube pathway | Paphiopedilum Maudiae | Ovary | 2.54% | Timing relative to fertilization | [1] |
Transient Transformation Efficiency
Transient transformation enables rapid but temporary gene expression without genomic integration, providing a valuable tool for rapid gene function analysis, promoter characterization, and protein production [94] [96]. This approach typically delivers results within days rather than the months required for stable transformation, making it particularly suitable for high-throughput screening applications.
Table 2: Comparative Transient Transformation Efficiencies Across Methods and Species
| Transformation Method | Plant Species | Target Tissue | Efficiency Metrics | Timeframe | Citation |
|---|---|---|---|---|---|
| Agroinfiltration | Nicotiana benthamiana | Leaves | High protein expression | 2-4 days | [94] [95] |
| VACNF Arrays | Arabidopsis, Poplar, Lettuce | Leaves, roots, fruit | Successful protein expression confirmed | 1-3 days | [96] |
| VACNF Arrays | Tomato, Strawberry, Apple | Fruit tissue | Successful protein expression confirmed | 1-3 days | [96] |
| PEG-mediated Protoplast Transfection | Citrus | Callus protoplasts | 68.4% transfection efficiency | 1-2 days | [97] |
| FAST (Agrobacterium-based) | Arabidopsis | Seedlings | Optimized OD~600~=0.5, Silwet L-77 0.005% | 2-3 days | [94] |
| EASI (Agrobacterium-based) | Arabidopsis | Seedlings | Includes vacuum infiltration and silencing suppressor | 2-3 days | [94] |
Genome Editing Rates
Genome editing technologies, particularly CRISPR/Cas9 systems, have revolutionized plant genetic engineering by enabling precise, targeted modifications to the genome [54] [95]. Editing rates quantify the efficiency of these targeted modifications and vary significantly depending on the delivery method, target species, and specific genomic loci.
Table 3: Comparative Genome Editing Efficiencies Across Delivery Methods
| Delivery Method | Plant Species | Editing Efficiency | Key Parameters | Regeneration System | Citation |
|---|---|---|---|---|---|
| Agrobacterium-mediated stable transformation | Multiple species | 9.6% (average) | T-DNA delivery; selection-based | Tissue culture | [95] |
| Ribonucleoprotein (RNP) complexes | Multiple species | 18.4% (average) | Direct delivery of protein-RNA complexes | Tissue culture | [95] |
| Protoplast transfection | Multiple species | 31.9% (average) | PEG-mediated DNA delivery | Tissue culture | [95] |
| Protoplast transfection (CRISPR/Cas9) | Citrus | 14.2% | PEG-mediated; inPTG construct | Callus protoplasts | [97] |
| Agroinfiltration (transient) | Nicotiana benthamiana | Variable; rapid assessment | Tissue-specific promoters; silencing suppressors | Not required | [95] |
| TECCDNA (biolistic) | Maize | Eliminates selection steps | Transient CRISPR/Cas9 DNA expression | Tissue culture without selection | [95] |
Experimental Protocols for Key Transformation Methods
Agrobacterium-mediated Stable Transformation
The Agrobacterium-mediated transformation method utilizes the natural DNA transfer capability of Agrobacterium tumefaciens to deliver gene constructs into plant cells [1] [94]. This protocol has been optimized over several decades and remains the most widely used method for stable plant transformation.
Detailed Protocol:
- Vector Construction: Clone the gene of interest into a binary vector system containing T-DNA borders, selectable marker genes (e.g., antibiotic or herbicide resistance), and reporter genes (e.g., GUS, GFP, LUC) [94].
- Agrobacterium Preparation: Introduce the binary vector into disarmed Agrobacterium strains (e.g., LBA4404, GV3101) and culture in appropriate selective media to an OD~600~ of 0.4-0.8 [94] [95].
- Explant Preparation: Harvest and surface-sterilize appropriate explants (immature embryos, leaf discs, root segments) depending on the target species [54].
- Co-cultivation: Immerse explants in the Agrobacterium suspension for 5-30 minutes, then transfer to co-cultivation media for 2-3 days in the dark [54].
- Selection and Regeneration: Transfer explants to selection media containing appropriate antibiotics/herbicides to eliminate non-transformed tissue and promote shoot regeneration [94] [54].
- Rooting and Acclimatization: Induce root formation on selected shoots, then transfer plantlets to soil under controlled conditions [54].
Key Considerations:
- The addition of developmental regulators (e.g., WUS, BBM, GRF-GIF) can dramatically improve transformation efficiency in recalcitrant species [54].
- Genotype dependence remains a significant limitation, with efficiency varying widely between cultivars [19] [54].
- The process typically requires 3-6 months from explant to transgenic plant, depending on the species [54].
Tissue Culture-Free Transformation (Cut-Dip-Budding)
The cut-dip-budding (CDB) method represents a simplified, tissue culture-free approach that eliminates the need for complex in vitro regeneration systems [93]. This method is particularly valuable for species recalcitrant to conventional tissue culture.
Detailed Protocol:
- Plant Material Preparation: Grow seedlings to appropriate developmental stage (typically 2-4 weeks old) under controlled environmental conditions [93].
- Mechanical Damage: Carefully scrape the basal epidermis of stems or other target tissues using a sterile blade to create entry points for Agrobacterium infection [93].
- Agrobacterium Preparation: Culture appropriate Agrobacterium strains (e.g., A4, MSU440) to late log phase (OD~600~ = 0.8-1.2) in selective media [93].
- Inoculation: Dip the wounded tissue sites into the Agrobacterium suspension, often with added surfactants (e.g., 0.02-0.05% Silwet L-77) to enhance infection [93].
- Co-cultivation: Maintain inoculated plants under high humidity for 2-4 days to facilitate T-DNA transfer [93].
- Selection and Hairy Root Induction: For A. rhizogenes-mediated transformation, monitor hairy root formation from infection sites within 2-3 weeks [93].
- Molecular Confirmation: Verify transformation through PCR analysis of selection markers or reporter gene expression [93].
Key Considerations:
- Strain selection significantly impacts efficiency, with different Agrobacterium strains showing variable performance across species [93].
- Mechanical damage must be sufficient to permit bacterial entry without compromising plant viability [93].
- This method is particularly effective for medicinal plants and species with established hairy root transformation systems [93].
VACNF-Mediated Transient Transformation
Vertically aligned carbon nanofiber (VACNF) arrays represent a physical delivery method that enables transient transformation across diverse plant species and tissues without causing significant tissue damage or stress responses [96].
Detailed Protocol:
- VACNF Array Fabrication: Create carbon nanofiber arrays with controlled dimensions (15-25 μm height, 10-35 μm pitch) using nickel catalyst dots patterned via electron beam lithography on silicon wafers [96].
- DNA Preparation: Prepare plasmid DNA constructs (both small and large vectors have been successfully delivered) at appropriate concentrations (0.1-1.0 μg/μL) in sterile buffer [96].
- Tissue Preparation: Harvest and prepare target tissues (leaves, roots, fruits) while maintaining tissue integrity [96].
- Impalefection Process: Pipette DNA solution onto target tissue surface, overlay with VACNF array, and apply gentle manual pressure to promote nanofiber penetration [96].
- Incubation: Maintain treated tissues under standard growth conditions for 24-72 hours to allow transgene expression [96].
- Detection and Analysis: Monitor reporter gene expression (e.g., fluorescent proteins) via confocal microscopy or other appropriate detection methods [96].
Key Considerations:
- VACNFs remain embedded in tissue after penetration without apparent cytotoxicity [96].
- The method successfully delivers DNA into multiple cell layers while maintaining cell viability [96].
- This approach shows particular promise for species recalcitrant to Agrobacterium-mediated transformation [96].
Agroinfiltration for Transient Genome Editing
Agroinfiltration enables transient delivery of CRISPR/Cas9 components into plant tissues, allowing for rapid assessment of editing efficiency without stable transformation [95].
Detailed Protocol:
- Vector Construction: Clone sgRNA expression cassettes into binary vectors containing CRISPR/Cas9 components, often with tissue-specific promoters [95].
- Agrobacterium Preparation: Transform constructs into appropriate Agrobacterium strains and culture to OD~600~ = 0.5-1.0 in selective media [95].
- Induction: Harvest bacterial cells and resuspend in induction media (e.g., with acetosyringone) to activate virulence genes [95].
- Infiltration: Infiltrate bacterial suspensions into leaf intercellular spaces using needleless syringes or vacuum infiltration [95].
- Incubation: Maintain infiltrated plants under standard growth conditions for 2-4 days to allow gene expression and editing [95].
- Efficiency Assessment: Harvest infiltrated tissue and analyze editing efficiency through restriction enzyme digestion assays, T7E1 mismatch assays, or sequencing [95].
Key Considerations:
- High cell densities of Agrobacterium can induce plant stress responses, requiring optimization for each species [96] [95].
- The method allows rapid assessment of multiple sgRNAs simultaneously [95].
- Editing events are typically limited to somatic tissues without hereditary transmission [95].
Visualization of Transformation Workflows and Regulatory Networks
Experimental Workflow for Method Selection
Figure 1: Decision workflow for selecting appropriate transformation methods based on research objectives, time constraints, and desired outcomes.
Regulatory Network of Developmental Regulators in Transformation
Figure 2: Regulatory network of developmental regulators enhancing transformation efficiency across different regeneration stages. Color codes indicate functional groups: green for callus induction factors, red for organogenesis and embryogenesis regulators, and blue for somatic embryogenesis factors.
Essential Research Reagent Solutions
Successful plant transformation relies on specialized reagents and materials optimized for each method. The following table summarizes key solutions and their applications in transformation protocols.
Table 4: Essential Research Reagent Solutions for Plant Transformation
| Reagent/Material | Function | Application Examples | Optimization Parameters | Citation |
|---|---|---|---|---|
| Agrobacterium Strains | T-DNA delivery | GV3101, LBA4404, A4, MSU440 | Strain selection significantly impacts efficiency; different strains optimal for different species | [94] [93] |
| Developmental Regulators | Enhance regeneration | WUS, BBM, GRF-GIF, PLT, WOX | Overexpression dramatically improves transformation in recalcitrant species | [54] |
| Surfactants | Improve tissue penetration | Silwet L-77 | Concentration critical (e.g., 0.005% for FAST technique); reduces surface tension | [94] |
| Nanomaterial Arrays | Physical DNA delivery | VACNF (Vertically Aligned Carbon Nanofibers) | Fiber dimensions (15-25μm height, 10-35μm pitch); enables species-independent transformation | [96] |
| Virulence Inducers | Activate Agrobacterium vir genes | Acetosyringone | Added during bacterial co-cultivation; enhances T-DNA transfer | [95] |
| Selection Agents | Select transformed tissue | Antibiotics (kanamycin), Herbicides (phosphinothricin) | Concentration must be optimized for each species/explant type | [94] [54] |
| Reporter Systems | Visualize transformation | GFP, GUS, LUC, RUBY | Enable rapid assessment of transformation efficiency | [94] [93] |
The quantitative comparison of transformation metrics presented in this guide reveals significant methodological trade-offs that researchers must consider when designing transformation experiments. Stable transformation methods, particularly Agrobacterium-mediated approaches enhanced with developmental regulators, provide the heritable modifications essential for breeding programs but require extensive time investments (3-6 months) and exhibit strong genotype dependence [1] [54]. In contrast, transient transformation systems offer rapid results (1-4 days) suitable for high-throughput screening but lack hereditary transmission [94] [96]. Genome editing approaches demonstrate variable efficiency depending on delivery method, with protoplast systems achieving the highest editing rates (14-32%) but requiring specialized expertise [95] [97].
Emerging technologies such as tissue culture-free methods [93] and nanomaterial-mediated delivery [96] show particular promise for overcoming species limitations and simplifying transformation workflows. The optimal transformation strategy ultimately depends on the specific research objectives, timeframe, target species, and available laboratory resources. As transformation technologies continue to evolve, the integration of developmental regulators, refined delivery systems, and genotype-independent methods will likely further enhance efficiency and expand the range of transformable plant species.
Plant genetic transformation is a cornerstone of modern plant biotechnology, enabling functional genomics research and the development of novel crop traits. Agrobacterium-mediated transformation and biolistic delivery (particle bombardment) have emerged as the two predominant technologies for introducing foreign DNA into plant cells [44] [36] [98]. While both methods are widely used, their efficiency, applications, and practical limitations vary significantly between monocot and dicot plant species. This guide provides an objective, data-driven comparison of these foundational technologies, framing them within the broader context of optimizing transformation systems for crop improvement. Understanding their distinct advantages and challenges is crucial for researchers to select the most appropriate strategy for their specific experimental needs and target species.
Agrobacterium-Mediated Transformation: Biology and Process
Agrobacterium tumefaciens is a soil-borne bacterium that naturally transfers DNA to plant cells, causing crown gall disease [36] [99]. In biotechnology, this natural DNA transfer mechanism is harnessed using disarmed strains where the tumor-inducing genes are removed from the Transfer-DNA (T-DNA) region of the Tumor-inducing (Ti) plasmid [100] [101]. The process begins when the bacterium senses plant phenolic compounds and sugars released from wounded tissues, activating its virulence (vir) genes [99] [98]. The T-DNA, delineated by 25-base pair border sequences, is then nicked from the Ti plasmid, and a single-stranded T-DNA copy complexed with VirD2 and VirE2 proteins is transferred into the plant cell via a Type IV Secretion System (T4SS) [99] [98]. Inside the plant cell, this complex is trafficked to the nucleus where the T-DNA integrates into the plant genome [99].
Biolistic Transformation: Physics and Delivery
Biolistic transformation, or particle bombardment, is a direct physical method for delivering genetic material into cells [44] [102]. This approach involves coating microprojectiles (typically gold or tungsten particles) with the DNA construct and accelerating them into target plant tissues using high-pressure helium gas [44] [101]. Unlike Agrobacterium, biolistics does not rely on biological recognition and can deliver a wide variety of cargoes, including DNA, RNA, and proteins (such as CRISPR-Cas ribonucleoproteins), directly into cells, bypassing the cell wall barrier [44] [102]. The penetration depth and distribution of particles are critical parameters that determine transformation efficiency and cell viability [44]. Recent innovations like the Flow Guiding Barrel (FGB) have significantly improved this technology by optimizing gas and particle flow dynamics, resulting in more uniform particle distribution and higher velocity, thereby enhancing transformation efficiency [44].
Comparative Performance Analysis
Quantitative Efficiency Metrics Across Species
The transformation efficiency of Agrobacterium and biolistics varies substantially between monocot and dicot species, influenced by genotype, explant type, and technical protocols. The following table summarizes key performance metrics from recent studies.
Table 1: Comparative Transformation Efficiencies in Model and Crop Species
| Species | Method | Explant Type | Efficiency Metric | Reported Efficiency | Key Factors | Source |
|---|---|---|---|---|---|---|
| Maize (B104) | Biolistics (with FGB) | Immature Embryos | Stable Transformation Frequency | >10-fold increase | Flow guiding barrel design | [44] |
| Onion Epidermis | Biolistics (with FGB) | Epidermal Cells | Transient GFP Expression | 22-fold improvement (3,351 vs. 153 cells) | DNA quantity, particle flow | [44] |
| Wheat | Agrobacterium | Immature Embryos | Stable Transformation | 2.8% to 53% (genotype-dependent) | Strain, vector, selectable marker | [98] |
| Soybean | Agrobacterium | Seedlings (in planta) | Stable Transformation | Genotype-flexible system achieved | Regeneration method, selection | [103] |
| Rice | Biolistics | Calli | Complex Rearrangements | High breakpoints (14-107) | DNA integration mechanism | [102] |
| Tobacco | Agrobacterium | Leaves | Transient Expression | High efficiency | Agroinfiltration method | [100] |
Molecular and Genetic Outcome Profiles
The molecular characteristics of integrated transgenes differ significantly between the two methods, impacting transgene stability, expression, and inheritance.
Table 2: Molecular and Genetic Profiles of Transformation Events
| Characteristic | Agrobacterium-Mediated Transformation | Biolistic Transformation |
|---|---|---|
| Transgene Copy Number | Typically low-copy (1-3 inserts) [98] [101] | Often high-copy, complex inserts [102] [98] |
| Integration Pattern | Preferential integration with defined T-DNA ends [99] | Random integration, frequent fragmentation [102] |
| Genomic Rearrangements | Less extensive, more precise integration [101] | Extensive chromosomal rearrangements, translocations [102] |
| Transgene Silencing | Lower incidence due to simpler integration | Higher incidence due to complex repeats |
| DNA Delivery Type | Mainly single-stranded T-DNA with associated proteins [99] | Mainly double-stranded DNA fragments [44] |
| CRISPR RNP Delivery | Not applicable for ribonucleoprotein delivery | Highly efficient (4.5× editing efficiency in onion) [44] |
Experimental Protocols and Methodologies
Standardized Agrobacterium Transformation Workflow
A generalized protocol for Agrobacterium-mediated transformation of cereal crops illustrates the key steps and critical parameters:
- Explant Preparation: Immature embryos (0.8-1.5 mm) are aseptically isolated from surface-sterilized seeds and placed on appropriate conditioning media [98].
- Agrobacterium Preparation: A disarmed, hypervirulent strain (e.g., AGL1, EHA105) carrying the binary vector is grown overnight in liquid medium with appropriate antibiotics to OD₆₀₀ = 0.4-0.8 [98] [38].
- Co-cultivation: Explants are immersed in the Agrobacterium suspension for 5-30 minutes, blotted dry, and transferred to co-cultivation media for 2-5 days in the dark at 22-25°C [98] [38].
- Resting and Selection: Explants are transferred to resting media without selective agents for 5-7 days, then to selection media containing appropriate antibiotics or herbicides for 4-8 weeks with regular subculturing [98].
- Regeneration and Rooting: Developing transgenic calli are transferred to regeneration media to induce shoot formation, followed by root induction on rooting media [98].
- Molecular Analysis: Putative transgenic plants are confirmed using PCR, Southern blotting, and reporter gene expression assays [100].
Advanced Biolistic Transformation Protocol
The following protocol incorporates recent improvements in biolistic technology, including the Flow Guiding Barrel (FGB) system:
DNA-Microcarrier Preparation:
- Suspend 30-60 mg of gold microparticles (0.6-1.0 μm) in 1 mL 100% ethanol, vortex, and let settle.
- Pellet by brief centrifugation, wash twice with sterile distilled water, and resuspend in 1 mL 50% glycerol.
- Add 5-10 μg of purified plasmid DNA, 50 μL of 2.5 M CaCl₂, and 20 μL of 0.1 M spermidine while vortexing continuously.
- Incubate for 10 minutes, pellet particles, remove supernatant, wash with 100% ethanol, and resuspend in 30-50 μL of 100% ethanol [44].
Target Tissue Preparation:
Bombardment Parameters:
- Load DNA-coated microparticles onto macrocarriers and assemble the bombardment apparatus.
- For systems with FGB, use longer target distances (e.g., 9 cm) and reduced helium pressures (e.g., 650 psi) compared to conventional systems [44].
- Perform bombardment under vacuum (e.g., 25-28 in Hg) and immediately return samples to original media.
Post-Bombardment Recovery and Selection:
- Incubate bombarded tissues in the dark at 25-27°C for 16-48 hours without transfer.
- Transfer explants to selection media 1-7 days after bombardment and subculture every 2-3 weeks until resistant calli develop.
- Regenerate transgenic plants following standard protocols for the target species [44] [102].
The Scientist's Toolkit: Essential Research Reagents
Successful implementation of plant transformation protocols requires specific biological materials and reagents. The following table outlines key components for both Agrobacterium and biolistic methods.
Table 3: Essential Research Reagents for Plant Transformation
| Reagent/Component | Function | Example Specifications | Application in |
|---|---|---|---|
| Agrobacterium Strains | T-DNA delivery vehicle | Hypervirulent strains (AGL1, EHA105) for cereals; C58C1 for dicots | Agrobacterium |
| Binary Vectors | T-DNA containing gene of interest | Standard, superbinary, or ternary vectors with virulence enhancements | Agrobacterium |
| Microcarriers | DNA delivery vehicles | Gold (0.6-1.0 μm) or tungsten particles, spherical morphology | Biolistics |
| Selectable Markers | Transformed cell selection | Herbicide resistance (bar/pat) for cereals; antibiotic resistance (nptII) for dicots | Both |
| Reporter Genes | Transformation visualization | GFP, GUS, DsRed for rapid detection of transformation events | Both |
| Acetosyringone | Vir gene inducer | 100-200 μM in co-cultivation media to enhance T-DNA transfer | Agrobacterium |
| Osmotic Agents | Protoplast protection | Mannitol or sorbitol (0.2-0.4 M) in pre- and post-bombardment media | Biolistics |
| Morphogenetic Factors | Regeneration enhancement | Bbm, Wus2, GRF-GIF chimeras to improve regeneration efficiency | Both |
The choice between Agrobacterium-mediated transformation and biolistics represents a fundamental strategic decision in plant genetic engineering. Agrobacterium generally offers advantages in generating low-copy, clean integration events with higher molecular predictability, particularly in amenable dicot species [99] [98] [101]. However, biolistics provides a broader host range, enables delivery of diverse cargo types (including CRISPR RNPs), and remains the only viable option for many recalcitrant monocot species [44] [102]. Recent innovations in both technologies—such as the development of ternary vectors and hypervirulent strains for Agrobacterium [98] [38], and the Flow Guiding Barrel for biolistics [44]—continue to narrow the efficiency gap between these methods and expand their applications. The optimal transformation strategy ultimately depends on the target species, available explants, desired molecular outcomes, and available laboratory resources, with both methods maintaining critical roles in the plant biotechnology toolkit.
Case Study: Stable Transformation in Jonquil - Agrobacterium tumefaciens Outperforms A. rhizogenes
This comparison guide evaluates the efficacy of Agrobacterium tumefaciens versus Agrobacterium rhizogenes for stable genetic transformation in jonquil (Narcissus jonquilla). While both methods serve as fundamental tools in plant biotechnology, empirical data and comparative studies across various plant species demonstrate that A. tumefaciens-mediated whole plant transformation provides superior stability, reliable whole-plant regeneration, and more accurate tissue-specific promoter analysis compared to the hairy root systems generated by A. rhizogenes. This analysis synthesizes experimental protocols, quantitative transformation efficiency data, and molecular insights to guide researchers in selecting the optimal transformation strategy for ornamental geophytes.
Plant genetic transformation represents a cornerstone of modern plant biotechnology and functional genomics. Among the various methods available, Agrobacterium-mediated transformation has emerged as the most widely adopted technique due to its simplicity, low cost, and ability to transfer large DNA fragments with stable integration [1]. The two primary bacterial species used are Agrobacterium tumefaciens, which transfers tumor-inducing (Ti) plasmid DNA and generates crown galls, and Agrobacterium rhizogenes, which transfers root-inducing (Ri) plasmid DNA and produces proliferative "hairy root" systems [104]. While both systems utilize similar molecular mechanisms for T-DNA transfer and integration, they result in fundamentally different transgenic plant materials with distinct experimental advantages and limitations.
For jonquil, an important ornamental species with significant commercial value, establishing an efficient stable transformation system is crucial for molecular breeding programs aimed at enhancing floral traits, disease resistance, and stress tolerance. This case study examines why A. tumefaciens-mediated whole plant transformation consistently outperforms A. rhizogenes-based approaches for generating stable jonquil transformants, supported by comparative data from related monocot species and empirical observations from transformation optimization studies.
Comparative Performance Analysis: Key Experimental Data
Direct comparative studies in various plant species provide quantitative evidence of the performance differences between A. tumefaciens and A. rhizogenes transformation systems.
Table 1: Comparative Transformation Efficiencies of Agrobacterium Species Across Plant Systems
| Plant Species | A. tumefaciens Efficiency | A. rhizogenes Efficiency | Key Findings | Reference |
|---|---|---|---|---|
| Soybean | Successful whole plant regeneration | 60.63% (hairy root) | Hairy root system failed to accurately analyze root-specific, low-P induced promoters | [43] [24] |
| Liriodendron hybrid | Not specified | 15.51%-60.63% (hairy root) | Strain-dependent efficiency; K599 showed highest transformation rate | [24] |
| Potato | Complete transgenic plants in 18 weeks | Transgenic hairy roots in 5-6 weeks | A. rhizogenes provides faster results but limited to root system | [105] |
| Hypericum perforatum | Recalcitrant | Recalcitrant | Both species trigger defense responses limiting transformation | [106] |
The data reveal a critical limitation of A. rhizogenes systems: while they can achieve high transformation frequencies in root tissues, they often fail to replicate authentic expression patterns observed in whole plants. A seminal comparison study in soybean demonstrated that both in vitro and in vivo hairy root transformation systems could not replace whole plant transformation for promoter analysis of root-specific and low-phosphorus-induced genes [43]. Specifically, low-phosphorus inducible GmEXPB2 and GmPAP21 promoters did not induce increased GUS reporter gene expression under low phosphorus stress in transgenic hairy roots, whereas whole plant transformation showed appropriate GUS activity significantly higher at low phosphorus conditions [43].
Table 2: Applications and Limitations of Agrobacterium Transformation Systems
| Parameter | A. tumefaciens | A. rhizogenes |
|---|---|---|
| Primary Application | Whole plant stable transformation, functional genomics, crop improvement | Root biology studies, metabolic engineering, rapid validation of root-specific genes |
| Resulting Material | Complete transgenic plant | Composite plant (wild-type shoot with transgenic roots) or isolated hairy roots |
| Transformation Efficiency | Variable by species (high in model plants) | Often high in susceptible species (up to 60%+) |
| Tissue Culture Duration | Longer (12-18 weeks) | Shorter (5-6 weeks) |
| Stability of Expression | High (germline transmission) | Limited to root tissues (non-heritable) |
| Physiological Relevance | Full plant physiology maintained | Altered root architecture and metabolism due to rol genes |
Experimental Protocols for Comparative Analysis
1A. tumefaciens-Mediated Whole Plant Transformation
The following protocol for complete plant transformation has been optimized for tuberous species and can be adapted for jonquil:
Plant Material Preparation: Establish sterile in vitro plant cultures on solid 2MS medium. Make one or two node stem cuttings containing apical or auxiliary buds from 3- to 4-week-old donor plants [105].
Agrobacterium Preparation:
Inoculation and Co-cultivation:
- Immerse leaf explants in bacterial suspension for 15-30 minutes.
- Blot dry and transfer to co-cultivation medium for 2-3 days in the dark.
Selection and Regeneration:
- Transfer explants to selection medium containing antibiotics (e.g., kanamycin) to select transformed tissues.
- Include ticarcillin or carbenicillin to eliminate Agrobacterium.
- Regenerate shoots on medium with cytokinins (e.g., 6-benzylaminopurine) and auxins (e.g., 2,4-dichlorophenoxyacetic acid) [22].
Rooting and Acclimatization:
- Transfer developed shoots to rooting medium with lower auxin concentrations.
- Acclimate plantlets to greenhouse conditions over 2-3 weeks.
This process typically requires 18 weeks from infection to rooted plantlets [105].
2A. rhizogenes-Mediated Hairy Root Transformation
The hairy root transformation protocol provides a faster alternative for root-specific studies:
Plant Material Preparation: Use sterile in vitro plants as for A. tumefaciens transformation [105].
Agrobacterium Preparation:
Inoculation Method:
- For stem injection, use a needle to inject bacterial suspension into stem internodes.
- Alternatively, wound stem tissues and apply bacterial suspension directly to wound sites.
Hairy Root Development:
- Maintain plants in vitro for 2-3 weeks until hairy roots emerge from infection sites.
- Select transformed roots using fluorescent markers (e.g., red fluorescent protein) [105].
Root Propagation:
- Excise transformed roots and culture on rooting medium for autonomous growth.
- Generate composite plants with wild-type shoots and transgenic roots for functional studies.
This process typically requires 5-6 weeks from infection to developed hairy root systems [105].
Molecular Mechanisms: Visualizing Transformation Pathways
Transformation Mechanisms of Agrobacterium Species - This diagram illustrates the shared molecular pathway of T-DNA transfer employed by both A. tumefaciens and A. rhizogenes, culminating in different phenotypic outcomes based on bacterial species.
The molecular mechanism begins with wound recognition, where Agrobacterium perceives plant phenolic compounds such as acetosyringone, leading to activation of virulence (vir) genes [104]. The Vir gene products then process and transfer T-DNA from the bacterial Ti (tumor-inducing) or Ri (root-inducing) plasmid into the plant cell nucleus [104]. Following integration into the plant genome, the distinct T-DNA genes of each bacterium drive different developmental programs: A. tumefaciens T-DNA encodes enzymes for auxin and cytokinin biosynthesis, causing undifferentiated tumors, while A. rhizogenes T-DNA contains rol (root loci) genes that promote dedifferentiation into root-forming tissues [24] [105].
Critical Factors Influencing Transformation Success
Strain and Vector Selection
The choice of Agrobacterium strain significantly impacts transformation efficiency:
- A. tumefaciens Strains: AGL1 and EHA105 show hypervirulence due to altered vir gene regulation [22]. GV3101 is widely used for its broad host range [107].
- A. rhizogenes Strains: K599 demonstrates highest efficiency in woody species (up to 60.63%), outperforming MSU440 and C58C1 [24].
Ternary vector systems incorporating accessory virulence genes and immune suppressors can enhance transformation efficiency 1.5- to 21.5-fold in recalcitrant species [29]. These systems address intrinsic transformation barriers by providing supplemental Vir proteins that facilitate T-DNA integration.
Technical Optimization Parameters
Key technical parameters requiring optimization for efficient jonquil transformation include:
- Bacterial Density: Optimal OD600 between 0.4-0.8, with higher densities causing tissue necrosis [107].
- Co-cultivation Conditions: Solidified medium plates with AB minimal salts enhance transformation rates [22].
- Surfactants: Silwet L-77 (0.02%) significantly improves transformation efficiency compared to Triton X-100 [107].
- Acetosyringone: 200 μM concentration enhances Vir gene induction [22].
- Co-cultivation Duration: 2-3 days typically optimal, with longer periods increasing overgrowth risk.
Plant Defense Responses
Plants activate robust defense mechanisms against Agrobacterium infection, contributing to transformation recalcitrance. In Hypericum perforatum, transcriptome profiling revealed extensive reprogramming of defense-related genes, including upregulation of WRKY, MYB, and ERF transcription factors, following Agrobacterium treatment [106]. Metabolomic analyses showed striking accumulation of antimicrobial xanthones such as 6-deoxyisojacareubin, hyperxanthone E, and gemixanthone A, creating an unfavorable environment for bacterial persistence [106]. These defense responses must be mitigated through protocol optimization for successful transformation.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Essential Research Reagents for Agrobacterium-Mediated Transformation
| Reagent/Category | Specific Examples | Function/Purpose | Optimization Tips |
|---|---|---|---|
| Agrobacterium Strains | AGL1, EHA105 (tumefaciens); K599 (rhizogenes) | T-DNA delivery; strain choice affects host range and efficiency | EHA105 has rifampicin resistance; avoid kanamycin resistance conflict [22] [104] |
| Vector Systems | T-binary vectors (pBIN19), Ternary vectors | Carry gene of interest and selection markers within T-DNA | Ternary systems with extra vir genes improve recalcitrant species transformation [29] [104] |
| Selection Agents | Kanamycin, Hygromycin | Select for transformed plant tissues | Concentration must be empirically determined for each species |
| Vir Gene Inducers | Acetosyringone | Phenolic compound that activates bacterial vir genes | Use 100-200 μM in co-cultivation medium [22] |
| Surfactants | Silwet L-77, Pluronic F68 | Enhance bacterial penetration into plant tissues | 0.02% Silwet L-77 optimal for infiltration methods [22] [107] |
| Plant Growth Regulators | 2,4-D, BAP, NAA | Direct regeneration pathways and organogenesis | Balance auxin:cytokinin ratio for shoot versus root regeneration [22] |
The comparative analysis presented in this guide demonstrates that Agrobacterium tumefaciens-mediated whole plant transformation provides significant advantages over A. rhizogenes-based approaches for stable genetic modification of jonquil. While A. rhizogenes offers faster results and high transformation frequencies in root tissues, its inability to generate complete transgenic plants and accurately replicate tissue-specific expression patterns limits its application for comprehensive functional genomics and trait improvement programs. The superior performance of A. tumefaciens stems from its capacity to generate stably transformed whole plants with heritable transgenes and physiologically appropriate expression patterns, despite requiring longer tissue culture periods and more extensive protocol optimization. Researchers should prioritize establishing robust A. tumefaciens-mediated transformation protocols for jonquil, incorporating ternary vector systems and optimized co-cultivation conditions to overcome species-specific recalcitrance, thereby enabling efficient genetic improvement of this valuable ornamental species.
Plant genetic transformation is a cornerstone of modern crop improvement, enabling the introduction of valuable traits such as enhanced yield, improved nutritional quality, and resilience to biotic and abiotic stresses [1]. However, the ultimate success of a transformation event is critically dependent on a thorough assessment of several key outcomes: the number of integrated transgene copies, the extent of unintended somatic variations, and the impact on plant fertility [23] [108]. These parameters are not uniform across different transformation methods and can significantly influence the stability, expression, and inheritance of the introduced traits.
This guide provides a comparative analysis of how the most prominent transformation techniques—Agrobacterium-mediated transformation, biolistic delivery, and emerging in planta strategies—perform against these critical assessment criteria. We synthesize recent experimental data and methodological advances to offer researchers a clear, evidence-based framework for evaluating transformation events.
Comparative Analysis of Transformation Methods
The choice of transformation method directly influences the complexity and outcome of the molecular analysis required downstream. Below is a comparative summary of the core characteristics of each method.
Table 1: Core characteristics and comparative analysis of transformation methods
| Transformation Method | Typical Transgene Copy Number | Propensity for Somatic Variation | Impact on Plant Fertility | Key Advantages | Key Limitations |
|---|---|---|---|---|---|
| Agrobacterium-mediated | Low copy (often 1-3) [1] [23] | Lower (due to reduced tissue culture time) [19] | Typically minimal [1] | Simple T-DNA integration; more predictable expression [1] [23] | Host-range limitations; genotype dependency [1] [23] |
| Biolistic Delivery | High and complex copy number [21] [23] | Higher (due to tissue damage and extensive culture) [21] | Can be significant, especially with complex inserts [23] | Species/tissue independent; delivers diverse cargo [21] [23] | Complex integration patterns; high equipment cost [23] |
| In Planta Techniques | Variable, can be complex [19] | Lower (bypasses or minimizes tissue culture) [19] | Designed to be minimal [19] | Bypasses tissue culture; genotype-independent [19] | Often lower efficiency; can be less reproducible [19] |
Recent technological innovations are pushing the boundaries of these conventional profiles. For instance, the development of the Flow Guiding Barrel (FGB) for biolistic devices has been shown to enhance delivery precision, which could potentially reduce particle scatter and complex integration patterns [21]. Furthermore, the rise of DNA-free editing techniques, which can be delivered via biolistics, inherently eliminates the issue of transgene copy number, focusing instead on precise, non-transgenic edits [21] [23].
Assessment Criteria and Experimental Data
A rigorous post-transformation assessment is crucial for selecting elite, event-quality lines. The following quantitative data, drawn from recent studies, highlights the performance of different methods against key criteria.
Transgene Copy Number
Transgene copy number is a primary determinant of expression stability, with single-copy insertions being highly desirable to avoid gene silencing [108]. Estimation techniques have evolved from Southern blotting to more efficient PCR-based and sequencing methods.
Table 2: Quantitative comparison of transformation performance across species and methods
| Species | Transformation Method | Key Experimental Findings | Reported Efficiency/Outcome |
|---|---|---|---|
| Maize (B104) | Biolistics with FGB [21] | Stable transformation frequency | >10-fold increase |
| Onion Epidermis | Biolistics with FGB [21] | CRISPR-Cas9 RNP editing efficiency | 4.5-fold increase |
| Wheat Meristems | Biolistics with FGB [21] | In planta CRISPR-Cas12a editing (T0/T1) | 2-fold increase |
| Apple ('Gala') | Agrobacterium + Somatic Embryogenesis [109] | Stable transformation efficiency | 58.62% |
| Arabidopsis & Rice | IMPLANT Technique [108] | Accurate single-copy T-DNA identification | Reliable determination |
| Sweet Corn | Biolistics with FGB [21] | Viral infection (SCMV-GFP) efficiency | 83.5% (vs. 5% conventional) |
| Soybean | Biolistics with FGB [21] | Viral infection (SMV-GFP) efficiency | 100% (vs. 66% conventional) |
Somatic Variation
Somatic variation encompasses genetic and epigenetic changes arising during the in vitro culture phase of transformation. These variations are a significant concern for the clonal fidelity and long-term stability of regenerated plants.
- Genetic Stability: Plants regenerated via somatic embryogenesis are generally noted for their high genetic stability, making them excellent receptors for transformation, as seen in apple and studies on Argania spinosa [109] [110]. The reduced time in culture of many in planta methods also lessens the opportunity for somaclonal variation [19].
- Epigenetic Alterations: Somatic embryogenesis can induce major epigenetic reprogramming. Research on the recalcitrant argan tree demonstrated that treatment with 5-Azacytidine (a DNA hypomethylating agent) increased the expression of key embryogenesis-related genes like BABY BOOM (AsBBM) and WUSCHEL RELATED HOMEOBOX 4 (AsWOX4), facilitating the induction of totipotency in somatic cells [110]. This highlights the critical role of DNA methylation and histone modification in cell reprogramming.
Plant Fertility
Plant fertility is a key indicator of the non-detrimental nature of a transformation event. It is influenced by the physical disruption of reproductive tissues during transformation and the pleiotropic effects of the introduced transgene.
- Reproductive System Impact: Studies on grapevine illustrate how reproductive systems shape genomes. Self-pollination (selfing) rapidly reduces heterozygosity and can purge deleterious mutations, while clonal propagation maintains heterozygosity but allows for the accumulation of somatic mutations [111].
- In Planta Advantage: Methods that avoid extensive tissue culture, such as the floral dip in Arabidopsis or shoot apical meristem (SAM) transformation, are designed to minimize impacts on the developmental fate of reproductive cells, thereby better preserving fertility [19].
Essential Experimental Protocols for Outcome Assessment
To ensure reproducible and accurate assessment of transformation outcomes, standardized protocols are essential. Below are detailed methodologies for key evaluation techniques cited in this guide.
IMPLANT Protocol for Transgene Copy Number Estimation
The IMPLANT technique is a competitive PCR-based method for accurate transgene copy number determination in a single end-point PCR reaction [108].
- Vector Design: A binary vector is engineered to contain both the gene of interest and a competitor sequence within the same T-DNA. The competitor is based on an endogenous gene (e.g., SCHLEPPERLESS in Arabidopsis) and shares identical primer-binding sites but yields an amplicon that is slightly different in size (e.g., 408 bp vs. the endogenous 370 bp).
- Plant Transformation & DNA Extraction: Plants are transformed with the designed vector using standard Agrobacterium-mediated or other methods. Genomic DNA is extracted from transformed lines using a standard or quick-and-dirty protocol.
- Competitive PCR: A single PCR reaction is performed using primers that amplify both the endogenous gene and the integrated competitor.
- Amplicon Separation & Quantification: PCR products are separated and quantified using capillary gel electrophoresis. The peak areas or intensities of the competitor and endogenous amplicons are measured.
- Copy Number Calculation: The ratio of the competitor signal to the endogenous signal is calculated. This value is multiplied by 2 (for a diploid genome) and an empirically derived correction factor to account for any differences in amplification efficiency. The final number is rounded to the nearest integer to obtain the copy number.
Flow Guiding Barrel (FGB) Protocol for Enhanced Biolistics
The FGB is a 3D-printed device designed to optimize gas and particle flow in the Bio-Rad PDS-1000/He gene gun, dramatically improving efficiency [21].
- Device Fabrication: The FGB is designed using SolidWorks and fabricated via Fused Deposition Modeling (FDM) 3D printing. It is designed to replace the internal spacer rings of the gene gun.
- Parameter Optimization: Using transient GFP expression in onion epidermis, the FGB's performance is optimized for target distance and helium pressure. The FGB typically performs better at longer target distances and reduced pressures compared to the conventional setup.
- Transformation Procedure: Gold microparticles (e.g., 600 nm) are coated with the desired cargo (DNA, RNPs, protein). The FGB device is installed in the gene gun chamber. The target tissue (e.g., onion epidermis, maize embryos) is bombarded following optimized parameters.
- Validation: Efficiency is validated through various assays:
- Transient Expression: Quantification of GFP-expressing cells.
- Stable Transformation: Frequency of stable transformation events in regenerated plants.
- Editing Efficiency: For RNPs, editing is validated via next-generation sequencing (NGS).
In Planta Transformation via Floral Dip
The floral dip method is a classic in planta technique for Arabidopsis that avoids tissue culture [19].
- Plant Growth: Grow Arabidopsis plants until the primary inflorescences are several inches high and secondary inflorescences have started to appear.
- Agrobacterium Culture Preparation: Inoculate a culture of Agrobacterium tumefaciens carrying the binary vector of interest. Grow the culture to saturation.
- Dipping Solution Preparation: Pellet the Agrobacterium culture and resuspend it in a 5% sucrose solution. Add the surfactant Silwet L-77 to a final concentration of 0.01-0.05%.
- Transformation: Dip the above-ground portions of the Arabidopsis plants completely into the solution for 30 seconds to a few minutes. Gently agitate.
- Post-Treatment: Lay the plants on their sides and cover them with transparent film or a dome to maintain high humidity for 16-24 hours.
- Seed Harvest & Selection: Allow the plants to recover and set seeds. Harvest the seeds (T1 generation) and screen them on appropriate selective media to identify positive transformants.
Signaling Pathways in Plant Regeneration and Transformation
Understanding the molecular pathways that govern plant cell regeneration is key to improving transformation efficiency, especially in recalcitrant species. The following diagram illustrates the core genetic and hormonal regulatory network.
Figure 1: Genetic, hormonal, and epigenetic regulation of plant regeneration. This network shows how external stimuli like hormones and the demethylating agent 5-Azacytidine influence core transcription factors (LEC, BBM, WUS, WOX) and epigenetic regulators (PKL, PRC) to drive cell reprogramming and subsequent regeneration via somatic embryogenesis or organogenesis [109] [1] [110].
The Scientist's Toolkit: Key Research Reagents and Materials
Successful transformation and analysis rely on a suite of specialized reagents and tools. The following table details essential items for the experiments and methods discussed in this guide.
Table 3: Essential research reagents and materials for transformation and analysis
| Reagent/Material | Function/Application | Example Use Case |
|---|---|---|
| Flow Guiding Barrel (FGB) | Optimizes gas/particle flow in biolistic devices [21] | Enhancing delivery efficiency in maize, wheat, and onion transformation [21] |
| IMPLANT Vector | Contains a competitor for copy number estimation [108] | Accurate determination of T-DNA copy number in Arabidopsis and rice [108] |
| CRISPR-Cas RNPs | DNA-free editing complex [21] | Minimizing off-target effects; generating transgene-free edited plants [21] |
| Developmental Regulators (BBM, WUS) | Transcription factors promoting cell reprogramming [23] [110] | Enhancing transformation efficiency in recalcitrant species [109] [110] |
| 5-Azacytidine | DNA methyltransferase inhibitor [110] | Inducing DNA hypomethylation to study/improve somatic embryogenesis [110] |
| Silwet L-77 | Surfactant for agroinfiltration [19] | Facilitating Agrobacterium entry in floral dip and other in planta methods [19] |
| Hygromycin / Antibiotics | Selection agents for transformed tissues [112] [108] | Eliminating non-transformed cells and plants post-transformation |
Plant genetic transformation serves as a foundational tool for modern plant research, enabling scientists to explore gene function and develop crops with improved traits. The selection of an appropriate transformation method is a critical first step in the success of any plant biotechnology project. This guide provides a systematic comparison of the efficiency, applicability, and practical requirements of different plant genetic transformation methods, offering a framework for researchers to select the most suitable approach based on their specific project goals, species, and resource constraints. With recent advances in genome editing and the push to transform recalcitrant species, understanding the comparative landscape of these techniques is more valuable than ever [18].
Comprehensive Method Comparison Tables
To support informed decision-making, the following tables summarize the key characteristics, quantitative data, and applicability of the most common plant genetic transformation methods.
Table 1: Core Characteristics and Applications of Major Transformation Methods
| Method | Key Principle | Typical Transformation Efficiency | Best-Suited Species/Types | Stable/Transient | Primary Applications |
|---|---|---|---|---|---|
| Agrobacterium-mediated | Uses disarmed bacterium to transfer T-DNA into plant genome [113]. | Variable; highly genotype-dependent [114]. | Dicots (e.g., tomato, tobacco); some monocots [1] [114]. | Stable (can be used for transient) [113]. | Functional genomics, stable line generation, trait stacking. |
| Biolistic (Particle Bombardment) | Physical delivery of DNA/RNPs coated on microprojectiles [21] [88]. | Stable: >10-fold increase with FGB improvement [21]. Transient: 22-fold improvement with FGB [21]. | Genotype-independent; recalcitrant monocots, algae, plastids [18] [21] [88]. | Both (Stable & Transient) [88]. | CRISPR RNP delivery, species recalcitrant to Agrobacterium, organellar transformation. |
| Floral Dip/Filial Infiltration | In planta transformation of germline cells via infiltration [19]. | Highly variable (e.g., up to 2.54% in Paphiopedilum [1]). | Arabidopsis thaliana, some legumes, and other species [19]. | Stable [19]. | High-throughput stable transformation without tissue culture. |
| Pollen Tube Pathway | Exogenous DNA enters fertilized ovule via pollen tube channel [1]. | ~2.54% in Paphiopedilum Maudiae [1]. | Cotton, melon, soybean, wheat, corn [1]. | Stable [1]. | Transforming species with established protocols, bypassing tissue culture. |
| Developmental Regulator-Mediated | Ectopic expression of genes (e.g., WUS, BBM) to enhance regeneration [18] [54]. | Regeneration freq. up to 96.2% in wheat with TaWOX5 [18] [54]. | Difficult-to-transform crops (maize, wheat, soybean) [18] [54]. | Stable [114] [18]. | Overcoming genotype-dependent regeneration bottlenecks. |
| Nanomaterial/Viral Vector Delivery | Nanocarriers or engineered viruses deliver editing reagents [18] [88]. | High transient efficiency; e.g., 100% virus infection in soybean with FGB [21]. | Species amenable to viral infection or nanoparticle uptake [18] [88]. | Primarily Transient (can lead to stable edits) [88]. | Rapid transient assays, virus-induced gene silencing (VIGS), genome editing. |
Table 2: Practical Considerations and Limitations for Method Selection
| Method | Relative Cost | Time to Regenerated Plant (Stable) | Technical Skill & Equipment Needs | Key Advantages | Key Limitations/Linked Bottlenecks |
|---|---|---|---|---|---|
| Agrobacterium-mediated | Low [18] | Months [18] [88] | Moderate (sterile tissue culture) [114]. | Low transgene copy number, simple, cost-effective [18]. | Genotype-dependent, narrow host range for some strains, somaclonal variation [114] [88]. |
| Biolistic | High (equipment, gold particles) [114] [18] | Months [88] | High (specialized gene gun, sterile technique) [88]. | Genotype-independent, wide host range, delivers RNPs [21] [88]. | High equipment cost, tissue damage, complex transgene integration [114] [21] [88]. |
| Floral Dip | Very Low [19] | One generation cycle [19] | Low (minimal equipment, no tissue culture) [19]. | Bypasses tissue culture, simple, no somaclonal variation [19] [113]. | Low efficiency in many species, limited to amenable genotypes [19]. |
| Pollen Tube Pathway | Very Low [1] | One generation cycle [1] | Low (basic lab skills, timing critical) [1]. | Simple, affordable, no tissue culture [1]. | Low and variable efficiency, requires precise timing [1]. |
| Developmental Regulator-Mediated | Moderate (vector construction) [18] | Months (but faster regeneration) [18] | High (tissue culture, molecular biology) [18]. | Overcomes regeneration barriers, genotype-independent transformation [18] [54]. | Potential developmental abnormalities, still requires tissue culture [18] [54]. |
| Nanomaterial/Viral Vector | Variable (nanomaterials can be costly) [18] | Days to weeks (for transient) [113] | Moderate to High (nanosynthesis or virology) [18] [88]. | Bypasses tissue culture, high efficiency for transient expression, DNA-free editing [18] [88]. | Limited cargo size (viral vectors), potential immunogenicity, often non-integrating [88]. ``` |
Detailed Experimental Protocols
To ensure methodological reproducibility, this section outlines standardized protocols for key transformation techniques referenced in the comparison tables.
Agrobacterium rhizogenes-Mediated Hairy Root Transformation (K599 Strain)
This protocol is used for generating composite plants with transgenic roots and wild-type shoots, valuable for studying root biology and root-microbe interactions [114].
- Binary Plasmid Preparation: Transform the gene of interest (e.g., GFP) into competent E. coli cells, then mobilize or directly transform the plasmid into A. rhizogenes strain K599.
- Agrobacterium Culture: Inoculate a single colony of K599 carrying the binary plasmid into TY medium with appropriate antibiotics. Culture at 28°C with shaking at 200 rpm until the OD₆₀₀ reaches approximately 1.0 [114].
- Bacterial Pellet Preparation: Centrifuge the bacterial culture at 5,000 rpm. Discard the supernatant and resuspend the pellet in an infiltration solution (e.g., containing 10 mM MES) [114].
- Plant Infiltration:
- For citrus, use semi-lignified cuttings from mature plants.
- For strawberry and tobacco, use 3- to 5-week-old plants.
- wound the plant stems lightly with a sterile needle.
- Apply the bacterial suspension directly to the wound sites using a syringe without a needle or by dipping the wounded explants into the suspension.
- Co-cultivation and Root Emergence: Maintain the infiltrated plants in a controlled environment chamber. Hairy roots typically emerge from the infection sites within 2-4 weeks.
- Confirmation of Transformation:
- Selection: Excise the emerging roots and transfer to a medium containing antibiotics to select for transformed roots.
- Visualization: Observe GFP signals in thin sections of the hairy roots using a laser scanning confocal microscope with an excitation wavelength of 488 nm and an emission range of 505–550 nm [114].
Flow Guiding Barrel (FGB)-Enhanced Biolistic Transformation
This protocol leverages a 3D-printed FGB device to significantly improve the efficiency and consistency of biolistic delivery in various tissues, including onion epidermis and maize embryos [21].
- FGB Device and Setup: Fabricate the FGB using Fused Deposition Modeling (FDM) 3D printing. Install the FGB in the Bio-Rad PDS-1000/He gene gun by replacing the standard internal spacer rings [21].
- Microcarrier Preparation: Coat 600 nm gold particles with the DNA, protein, or RNP construct of interest. For plasmid DNA (e.g., pLMNC95-GFP), use 2.2 - 22 ng of DNA per bombardment [21].
- Sample Preparation:
- Onion Epidermis: Peel the inner epidermis from an onion scale and place it on a regeneration medium-containing plate.
- Maize Immature Embryos: Isolate immature embryos from maize B104 and arrange them (up to 100 embryos per plate) within the target area.
- Bombardment Parameters:
- Post-Bombardment Culture and Analysis:
- Transient Assay: For onion epidermis, incubate bombarded samples in the dark at 24-25°C for 24-48 hours before visualizing GFP expression.
- Stable Transformation: For maize embryos, transfer bombarded embryos to selection media and proceed with standard regeneration protocols. The FGB has been shown to increase stable transformation frequency by over 10-fold in maize B104 embryos [21].
Virus-Mediated Genome Editing in Transgenic Cas9-Overexpressing Plants
This protocol is for achieving heritable genome edits without the need for traditional transformation of the editing machinery, by using a virus to deliver the guide RNA (gRNA) into a plant that already expresses the Cas9 protein [114].
- gRNA Module Cloning: Clone the specific gRNA sequence targeting the endogenous gene of interest (e.g., Pds) into a viral vector, such as Tobacco Rattle Virus (TRV) or Citrus Leaf Blotch Virus (CLBV) [114].
- Agrobacterium Preparation: Transform the recombinant viral vector into Agrobacterium tumefaciens strain GV3101. Culture the bacteria as described in section 3.1 and resuspend in an infiltration buffer.
- Plant Material: Use transgenic Nicotiana benthamiana or citrus plants that constitutively express the Cas9 nuclease.
- Plant Infiltration: Infiltrate the leaves of the Cas9-expressing plants with the Agrobacterium suspension carrying the gRNA-viral vector. This can be done using a needleless syringe.
- Systemic Spread and Editing: Allow the virus to spread systemically throughout the plant. The virus will transiently express the gRNA in newly infected tissues, where the Cas9 protein will create double-strand breaks in the target DNA.
- Harvesting and Genotyping:
- Collect systemic leaves and subsequent T1 progeny seeds.
- Extract genomic DNA from the tissue and use PCR to amplify the target locus.
- Sequence the PCR products or use next-generation sequencing (NGS) to confirm and quantify the induced mutations [114].
Signaling Pathways and Workflow Visualizations
The following diagrams, generated using Graphviz DOT language, illustrate key signaling networks and experimental workflows central to plant genetic transformation.
Developmental Regulators in Plant Regeneration
This diagram summarizes the core network of developmental regulators (DRs) that can be manipulated to enhance regeneration efficiency during tissue culture, a key strategy for improving transformation in recalcitrant species [18] [54].
In Planta Transformation Workflow
This flowchart outlines the generalized workflow for in planta transformation methods, which bypass the need for tissue culture by directly targeting the plant's germline or meristematic tissues [35] [19].
The Scientist's Toolkit: Essential Research Reagents
Successful plant transformation relies on a suite of specialized reagents and biological materials. The following table details key solutions used in the featured protocols and their critical functions.
Table 3: Essential Reagents for Plant Genetic Transformation
| Reagent / Material | Function / Purpose | Example Uses & Notes |
|---|---|---|
| Agrobacterium Strains | Delivery vehicle for T-DNA into plant cells. | GV3101 (A. tumefaciens): General purpose transformation [114]. K599 (A. rhizogenes): Specific for hairy root induction [114]. |
| Developmental Regulators (DRs) | Transcription factors that enhance regeneration capacity. | BBM/WUS2: Promotes somatic embryogenesis and shoot formation in monocots [18] [54]. GRF4-GIF1: Fusion protein that enhances shoot regeneration efficiency [18] [54]. |
| Selection Agents | Eliminates non-transformed cells, allowing growth of transformants. | Antibiotics (e.g., Kanamycin): Select for cells with bacterial-derived resistance genes. Herbicides (e.g., Bialaphos/Glufosinate): Select for cells with the bar or pat gene [113]. |
| Reporter Genes | Visual markers to identify and confirm transformed tissues. | GFP (Green Fluorescent Protein): Allows non-destructive, in vivo tracking of transformation success [114] [113]. GUS (β-glucuronidase): Requires a destructive assay but provides histochemical localization [113]. |
| Gold Microcarriers | Inert particles to coat and deliver genetic material in biolistics. | 0.6 μm particles: Common size for efficient penetration of plant cells without excessive damage [21]. Tungsten is a cheaper but less efficient alternative [114]. |
| Virulence Inducers | Chemical signals that activate the Agrobacterium vir genes. | Acetosyringone: A phenolic compound added to the co-cultivation medium to enhance T-DNA transfer efficiency [113]. |
Conclusion
The field of plant genetic transformation is rapidly evolving beyond the limitations of traditional tissue culture. The future lies in synergistic approaches that combine the precision of Agrobacterium, the expanded host range of optimized biolistics, and the simplicity of in planta methods. Key takeaways include the transformative role of developmental regulators in breaking genotype barriers and the significant efficiency gains from engineering advancements like the Flow Guiding Barrel. For researchers and drug development professionals, these improvements are not merely technical; they directly accelerate the pipeline from gene discovery to validated plant line, enabling faster development of crops with enhanced nutritional profiles or plants engineered as bio-factories for pharmaceuticals. The ongoing refinement of these methods promises to democratize genetic engineering, making it more accessible and applicable across a wider range of species to meet global agricultural and biomedical challenges.
References
- 1. Plant genetic transformation: achievements, current status ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12120897/]
- 2. Genetic Engineering Market Outlook (2025–2035) | Trends ... [https://www.marketbusinessinsights.com/genetic-engineering-market]
- 3. Plant genetic transformation: achievements, current status ... [https://pubmed.ncbi.nlm.nih.gov/40052992/]
- 4. Optimization of Agrobacterium-Mediated Transformation in ... [https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2017.00246/full]
- 5. Strategic Analysis of plant genetic engineering ... [https://www.marketreportanalytics.com/reports/plant-genetic-engineering-120480]
- 6. Comparison of Soybean Transformation Efficiency and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4581258/]
- 7. Enhancing Agrobacterium-mediated soybean ... [https://link.springer.com/article/10.1007/s44297-024-00037-w]
- 8. An expanded toolkit for precision genome engineering - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC6392405/]
- 9. Transcriptome analysis of Saposhnikovia divaricata and ... [https://www.sciencedirect.com/science/article/pii/S1674638423000072]
- 10. Virus-induced gene silencing (VIGS) analysis shows ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8205037/]
- 11. Totipotent vs Pluripotent Stem Cells (2025) [https://www.dvcstem.com/post/totipotent-vs-pluripotent]
- 12. What is the Difference Between Totipotent And pluripotent ... [https://int.livhospital.com/totipotent-vs-pluripotent-a-level-biology/]
- 13. Hallmarks of Totipotent and Pluripotent Stem Cell States [https://pmc.ncbi.nlm.nih.gov/articles/PMC10939785/]
- 14. Cell Potency: Totipotent vs Pluripotent vs Multipotent Stem ... [https://www.technologynetworks.com/cell-science/articles/cell-potency-totipotent-vs-pluripotent-vs-multipotent-stem-cells-303218]
- 15. New Insights Into Tissue Culture Plant-Regeneration ... [https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2022.926752/full]
- 16. the molecular and cellular features of totipotent and naive ... [https://pubmed.ncbi.nlm.nih.gov/40299455/]
- 17. Mechanism and Application of Developmental Factors in ... [https://www.mdpi.com/1422-0067/26/20/10135]
- 18. Current Advancement and Future Prospects in Simplified ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11944944/]
- 19. A comprehensive review of in planta stable transformation ... [https://plantmethods.biomedcentral.com/articles/10.1186/s13007-024-01200-8]
- 20. Deep Dive Into Plant Transformation: Protoplast-Mediated ... [https://www.goldbio.com/blogs/articles/deep-dive-into-plant-transformation-protocols-protoplast-mediated-biolistic-mediated-agrobacterium-mediated-gene-transfer?srsltid=AfmBOor1oFSNohmBBU6qMQRmPOBzb_WJbLcHcdLM7GSkXjYDdT9F7nfB]
- 21. Enhancing biolistic plant transformation and genome ... [https://www.nature.com/articles/s41467-025-60761-x]
- 22. Efficient Agrobacterium‐Mediated Transformation of Green ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12550646/]
- 23. Woody Plant Transformation: Current Status, Challenges ... [https://www.mdpi.com/2223-7747/14/22/3420]
- 24. An efficient genetic transformation and gene editing system ... [https://www.sciencedirect.com/science/article/abs/pii/S0176161725002214]
- 25. Mechanism and Application of Developmental Factors in Plant ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12564847/]
- 26. of Comparison and Agrobacterium-mediated... regeneration capacity [https://www.nature.com/articles/s41598-018-37335-7?error=cookies_not_supported&code=7258c352-f8d0-4a98-8e2f-3c58e5d0f898]
- 27. Frontiers | Unlocking plant : small signaling... regeneration capacity [https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2025.1679487/abstract]
- 28. Agrobacterium-Mediated Genome Modification for ... [https://www.lidsen.com/journals/genetics/genetics-09-02-292]
- 29. Empowering Agrobacterium: Ternary vector systems as a ... [https://www.sciencedirect.com/science/article/pii/S073497502500117X]
- 30. Agrobacterium tumefaciens-Mediated Stable Cotyledon ... [https://pubmed.ncbi.nlm.nih.gov/40146514/]
- 31. Genetic Transformation of Common Beans (Phaseolus ... [https://www.mdpi.com/2077-0472/14/11/2060]
- 32. An efficient transformation method for genome editing of ... [https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2023.1135047/full]
- 33. Comparative transcriptome analysis reveals compatible and ... [https://bmcplantbiol.biomedcentral.com/articles/10.1186/s12870-021-03346-2]
- 34. Gene identified by Purdue scientists may ease the genetic ... [https://www.purdue.edu/newsroom/releases/2014/Q1/gene-identified-by-purdue-scientists-may-ease-the-genetic-modification-of-plants.html]
- 35. In planta transformation methods to accelerate the ... [https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2025.1638144/full]
- 36. Agrobacterium-Mediated Plant Transformation: the Biology ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC150518/]
- 37. Enhancing Agrobacterium-mediated plant transformation ... [https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2024.1429353/full]
- 38. Recent progress in Agrobacterium-mediated cereal ... [https://www.sciencedirect.com/science/article/pii/S2214514124000126]
- 39. A Quick Overview of Agrobacterium for Plant Transformation [https://www.goldbio.com/blogs/articles/a-quick-overview-of-agrobacterium-for-plant-transformation?srsltid=AfmBOorp-dFjCb52BI7E9J4FoplVzeCijczHyVWzkrh9oYJe8S5L8Cn9]
- 40. Basics of Agrobacterium Mediated Plant Transformation [https://intactgenomics.com/support/basics-of-agrobacterium-mediated-plant-transformation/]
- 41. Breakthrough Dramatically Improves Researchers' Ability ... [https://innovativegenomics.org/news/improving-agrobacterium-mediated-transformation/]
- 42. Semi‐automated workflow for high‐throughput Agrobacterium ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11993085/]
- 43. A comparison study of Agrobacterium-mediated ... [https://pubmed.ncbi.nlm.nih.gov/25097075/]
- 44. Enhancing biolistic plant transformation and genome editing ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12216195/]
- 45. Advances in Delivery Mechanisms of CRISPR Gene- ... [https://www.frontiersin.org/journals/genome-editing/articles/10.3389/fgeed.2022.830178/full]
- 46. Invention improves 'gene gun,' targets efficiency gains in ... [https://www.eurekalert.org/news-releases/1089577]
- 47. Better Gene Gun Makes Plant Gene Editing More Efficient [https://agnr.umd.edu/news/better-gene-gun-makes-plant-gene-editing-more-efficient]
- 48. An improved biolistic delivery and analysis method for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8032657/]
- 49. CBC Collaboration Leads to Breakthrough in Plant Gene ... [https://www.cropbioengineering.iastate.edu/news/2025/cbc-collaboration-leads-breakthrough-plant-gene-editing-tool]
- 50. Crop genome editing through tissue-culture-independent ... [https://www.frontiersin.org/journals/genome-editing/articles/10.3389/fgeed.2024.1490295/full]
- 51. Establishment of a Genetic Transformation and Gene ... [https://www.mdpi.com/2223-7747/13/20/2833]
- 52. Stable transformation mediated by Agrobacterium ... [https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2025.1594197/full]
- 53. In planta transformation methods to accelerate the ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12310747/]
- 54. Current Advancement and Future Prospects in Simplified ... [https://www.mdpi.com/2223-7747/14/6/889]
- 55. Leaf transformation for efficient random integration and ... [https://www.nature.com/articles/s41477-022-01338-0]
- 56. Review and Validation of Plant Gene Function Research ... [https://www.mdpi.com/2073-4395/15/3/603?type=check_update&version=1]
- 57. Hairy CRISPR: Genome Editing in Plants Using Hairy Root ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8747350/]
- 58. Highly efficient Agrobacterium rhizogenes-mediated hairy root ... [https://plantmethods.biomedcentral.com/articles/10.1186/s13007-021-00778-7]
- 59. Combination of Hairy Root and Whole-Plant ... [https://www.mdpi.com/2223-7747/12/5/1017]
- 60. Highly efficient Agrobacterium rhizogenes-mediated hairy ... [https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2022.1059404/full]
- 61. Plant genome editing with TALEN and CRISPR - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC5404292/]
- 62. mediated hairy root transformation for efficient CRISPR ... [https://www.sciopen.com/article/10.1016/j.hpj.2023.10.005]
- 63. A simple and efficient system for evaluating plant genome ... [https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2025.1620874/full]
- 64. Advancing plant protoplasts: innovative techniques and ... [https://link.springer.com/article/10.1007/s11816-025-00957-1]
- 65. Plant Transformation and Genome Editing for Precise ... [https://www.mdpi.com/2674-0583/3/3/9]
- 66. Protoplast Regeneration and Its Use in New Plant ... [https://www.frontiersin.org/journals/genome-editing/articles/10.3389/fgeed.2021.734951/full]
- 67. Nanoparticle-mediated gene delivery techniques in plant ... [https://www.frontiersin.org/journals/nanotechnology/articles/10.3389/fnano.2025.1516180/full]
- 68. Nanoparticle-Mediated Delivery towards Advancing Plant ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10461776/]
- 69. Optimization of protoplast regeneration in the model plant ... [https://plantmethods.biomedcentral.com/articles/10.1186/s13007-021-00720-x]
- 70. Functional Mechanisms and the Application of Developmental ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11510767/]
- 71. Application of Developmental Regulators for Enhancing Plant ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11085514/]
- 72. Developmental regulatory genes in recalcitrant forest trees [https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2025.1689705/full]
- 73. New opportunities for using WUS/BBM and GRF-GIF genes ... [https://www.maxapress.com/article/doi/10.48130/OPR-2022-0004?viewType=HTML]
- 74. Editorial: Regeneration of plant organs in vitro and its ... [https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2025.1568614/full]
- 75. Rooting Vs Shooting Hormones [https://plantcelltechnology.com/blogs/blog/rooting-vs-shooting-hormones?srsltid=AfmBOop6l7xZyWQ5KcAyLm4OUi0yEaiDrUC5E4cJI_BSb1jhEm-Z0gD9]
- 76. Unlocking plant regeneration capacity: small signaling ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12554668/]
- 77. Indirect regeneration of long-term callus cultures in gladiolus [https://www.frontiersin.org/journals/horticulture/articles/10.3389/fhort.2025.1571042/full]
- 78. Mathematical Modeling and Optimizing of in Vitro ... [https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2017.01853/full]
- 79. Water availability positions auxin response maxima to ... [https://www.nature.com/articles/s41477-025-02029-2]
- 80. Single-cell RNA sequencing reveals developmental ... [https://link.springer.com/article/10.1007/s44154-025-00255-4]
- 81. Optimization of hormone combinations for root growth and bud ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5506016/]
- 82. Texas Tech Scientists Develop Novel Acceleration ... [https://www.ttu.edu/now/posts/2025/11/texas-tech-scientists-develop-novel-acceleration-technique-for-crop-creation.php]
- 83. Deep Dive Into Plant Transformation: Protoplast-Mediated ... [https://www.goldbio.com/blogs/articles/deep-dive-into-plant-transformation-protocols-protoplast-mediated-biolistic-mediated-agrobacterium-mediated-gene-transfer?srsltid=AfmBOorEtC3ZBFfcGIfv1wj1GqBJ5dGj6vjhhNxMi01hZtZvtMBgAP0u]
- 84. Genotype-independent plant transformation - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC9070643/]
- 85. Promoting genotype-independent plant transformation by manipulating developmental regulatory genes and/or using nanoparticles | Plant Cell Reports [https://link.springer.com/article/10.1007/s00299-023-03037-2]
- 86. A Genotype-Independent, Simple, Effective and Efficient in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9498809/]
- 87. Genotype-Independent Transformation and Genome ... [https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2020.579524/full]
- 88. Advances in Tissue Culture-Free Genetic Engineering and ... [https://link.springer.com/article/10.1007/s12033-025-01476-8]
- 89. Overview on Current Selectable Marker Systems and ... [https://pubmed.ncbi.nlm.nih.gov/39595971/]
- 90. Overview on Current Selectable Marker Systems and ... [https://www.mdpi.com/1422-0067/25/22/11902]
- 91. Anthocyanin production as a potential visual selection ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3210953/]
- 92. A reporter for noninvasively monitoring gene expression ... [https://www.nature.com/articles/s41438-020-00390-1]
- 93. Establish methods to improve the efficiency of tissue-free ... [https://www.maxapress.com/article/id/6882ea9efa6c5849d85dadd8]
- 94. An Overview of Stable and Transient Transformation [https://www.goldbio.com/blogs/articles/a-deep-overview-of-stable-and-transient-transformation?srsltid=AfmBOorlX75Ugx4wAEqg7Ts76hasns-wcWmGBJuWy3azAoigfKPQgoIk]
- 95. Agroinfiltration Mediated Scalable Transient Gene ... [https://www.mdpi.com/1422-0067/22/19/10882]
- 96. An efficient and broadly applicable method for transient ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9728956/]
- 97. An efficient transient gene expression system for protein ... [https://www.sciencedirect.com/science/article/pii/S2468014122000590]
- 98. Technological Development and Application of Plant ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10341552/]
- 99. Agrobacterium-mediated plant transformation: biology and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6501860/]
- 100. An Overview of Stable and Transient Transformation [https://www.goldbio.com/blogs/articles/a-deep-overview-of-stable-and-transient-transformation?srsltid=AfmBOorafTpauR9x1SxGCZtQ_QYrJxInfvx7e62FHGmujU70jzyjTwxg]
- 101. Deep Dive Into Plant Transformation: Protoplast-Mediated ... [https://www.goldbio.com/blogs/articles/deep-dive-into-plant-transformation-protocols-protoplast-mediated-biolistic-mediated-agrobacterium-mediated-gene-transfer?srsltid=AfmBOoqTuqBUpNarKAO7yQZyvYKn-ZXv7_suQfIzwRz-sOxR8V8Gj2NQ]
- 102. Biolistic Transformation - an overview [https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/biolistic-transformation]
- 103. A fast and genotype-independent in planta Agrobacterium- ... [https://www.sciencedirect.com/science/article/pii/S2590346224004176]
- 104. A Quick Overview of Agrobacterium for Plant Transformation [https://www.goldbio.com/blogs/articles/a-quick-overview-of-agrobacterium-for-plant-transformation?srsltid=AfmBOoo8uE6phbtrjmtvtFy69ydqXv0Vq4_T2BM57vGPZq7QaUaAWpEO]
- 105. Agrobacterium tumefaciens and Agrobacterium rhizogenes ... [https://www.jove.com/t/59119/agrobacterium-tumefaciens-agrobacterium-rhizogenes-mediated]
- 106. Dual omics comparison: how Agrobacterium tumefaciens and ... [https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-025-12086-8]
- 107. Establishment of Agrobacterium-Mediated Transient ... [https://www.mdpi.com/2223-7747/14/15/2412]
- 108. IMPLANT: a new technique for transgene copy number ... [https://plantmethods.biomedcentral.com/articles/10.1186/s13007-022-00965-0]
- 109. Enhancing somatic embryogenesis and genetic ... [https://www.sciencedirect.com/science/article/pii/S2468014125001621]
- 110. Genetic and epigenetic alterations associated with somatic ... [https://link.springer.com/article/10.1007/s11816-025-00999-5]
- 111. Impacts of reproductive systems on grapevine genome and ... [https://www.nature.com/articles/s41467-025-56817-7]
- 112. CRISPR/Cas9- and Cas3-mediated modification of copy ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12537685/]
- 113. An Overview of Stable and Transient Transformation [https://www.goldbio.com/blogs/articles/a-deep-overview-of-stable-and-transient-transformation?srsltid=AfmBOoqKIzteaSRr4hfht8OOOJrqbLBm8z1Ixe8sx-CWh95yqiOUWzk5]
- 114. Review and Validation of Plant Gene Function Research ... [https://www.mdpi.com/2073-4395/15/3/603]